Shumeng Li
Dual-Teacher Ensemble Models with Double-Copy-Paste for 3D Semi-Supervised Medical Image Segmentation
Oct 15, 2024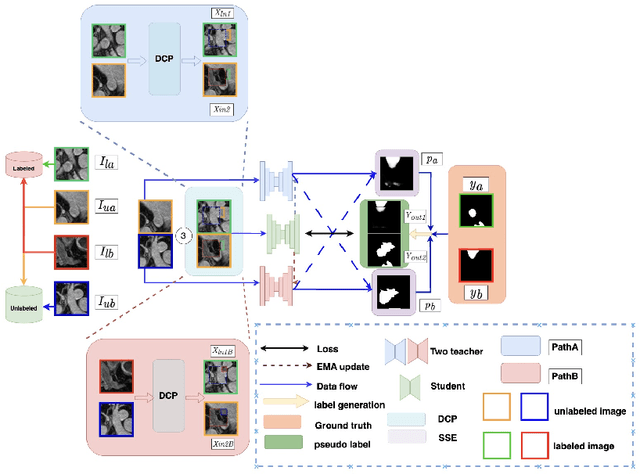
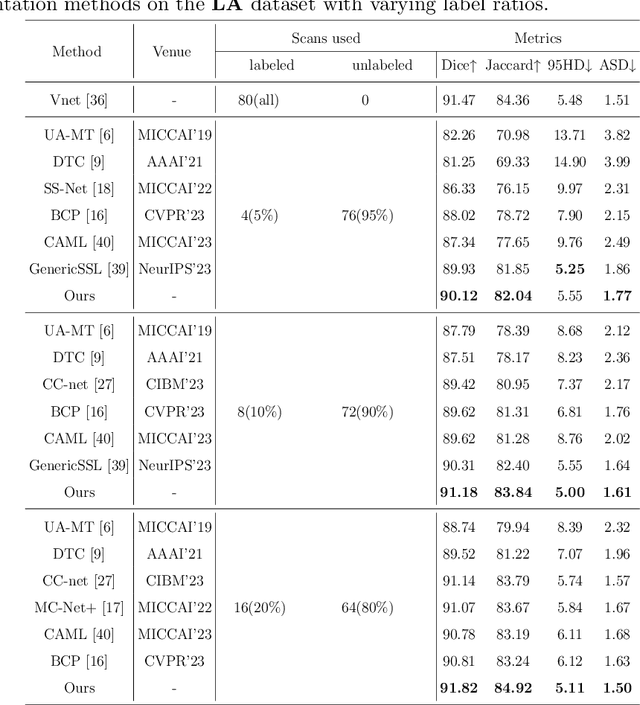
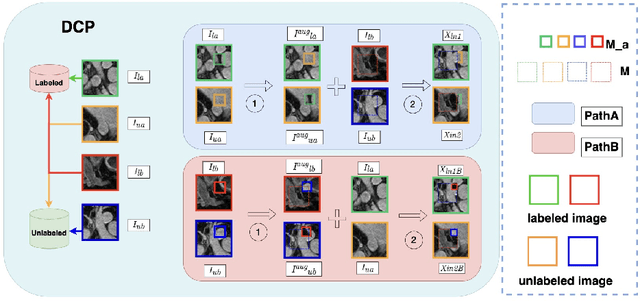
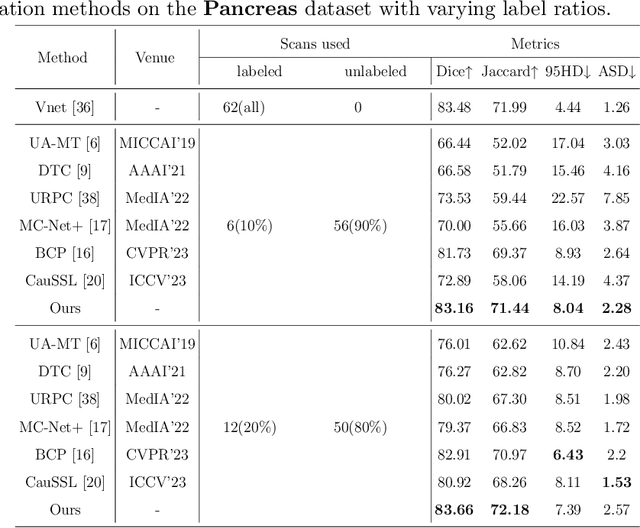
Abstract:Semi-supervised learning (SSL) techniques address the high labeling costs in 3D medical image segmentation, with the teacher-student model being a common approach. However, using an exponential moving average (EMA) in single-teacher models may cause coupling issues, where the weights of the student and teacher models become similar, limiting the teacher's ability to provide additional knowledge for the student. Dual-teacher models were introduced to address this problem but often neglected the importance of maintaining teacher model diversity, leading to coupling issues among teachers. To address the coupling issue, we incorporate a double-copy-paste (DCP) technique to enhance the diversity among the teachers. Additionally, we introduce the Staged Selective Ensemble (SSE) module, which selects different ensemble methods based on the characteristics of the samples and enables more accurate segmentation of label boundaries, thereby improving the quality of pseudo-labels. Experimental results demonstrate the effectiveness of our proposed method in 3D medical image segmentation tasks. Here is the code link: https://github.com/Fazhan-cs/DCP.
Concatenate, Fine-tuning, Re-training: A SAM-enabled Framework for Semi-supervised 3D Medical Image Segmentation
Mar 17, 2024Abstract:Segment Anything Model (SAM) fine-tuning has shown remarkable performance in medical image segmentation in a fully supervised manner, but requires precise annotations. To reduce the annotation cost and maintain satisfactory performance, in this work, we leverage the capabilities of SAM for establishing semi-supervised medical image segmentation models. Rethinking the requirements of effectiveness, efficiency, and compatibility, we propose a three-stage framework, i.e., Concatenate, Fine-tuning, and Re-training (CFR). The current fine-tuning approaches mostly involve 2D slice-wise fine-tuning that disregards the contextual information between adjacent slices. Our concatenation strategy mitigates the mismatch between natural and 3D medical images. The concatenated images are then used for fine-tuning SAM, providing robust initialization pseudo-labels. Afterwards, we train a 3D semi-supervised segmentation model while maintaining the same parameter size as the conventional segmenter such as V-Net. Our CFR framework is plug-and-play, and easily compatible with various popular semi-supervised methods. Extensive experiments validate that our CFR achieves significant improvements in both moderate annotation and scarce annotation across four datasets. In particular, CFR framework improves the Dice score of Mean Teacher from 29.68% to 74.40% with only one labeled data of LA dataset.
Orthogonal Annotation Benefits Barely-supervised Medical Image Segmentation
Mar 23, 2023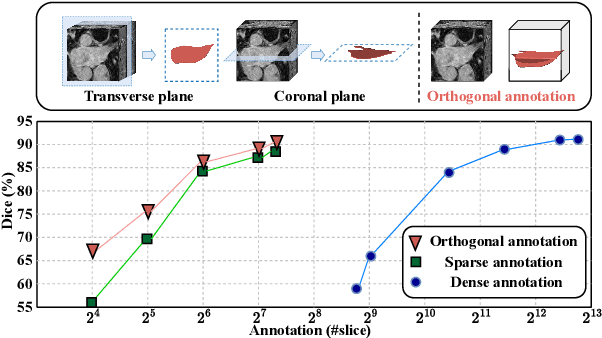
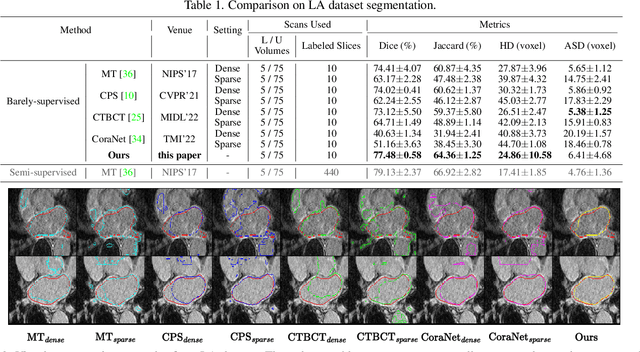
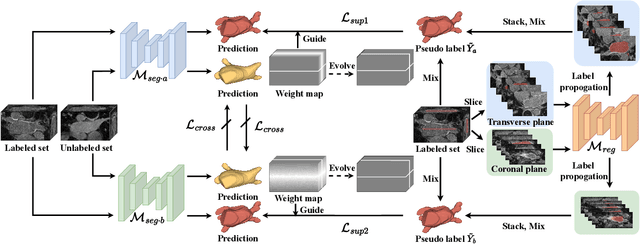
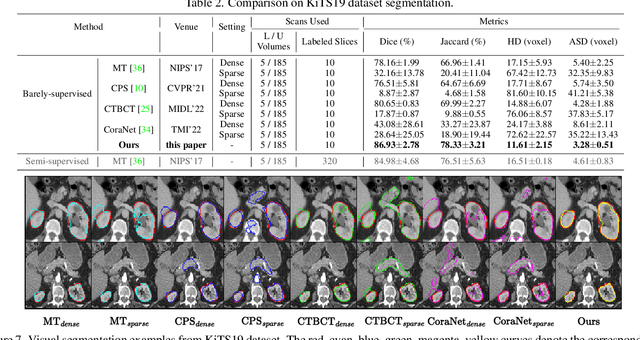
Abstract:Recent trends in semi-supervised learning have significantly boosted the performance of 3D semi-supervised medical image segmentation. Compared with 2D images, 3D medical volumes involve information from different directions, e.g., transverse, sagittal, and coronal planes, so as to naturally provide complementary views. These complementary views and the intrinsic similarity among adjacent 3D slices inspire us to develop a novel annotation way and its corresponding semi-supervised model for effective segmentation. Specifically, we firstly propose the orthogonal annotation by only labeling two orthogonal slices in a labeled volume, which significantly relieves the burden of annotation. Then, we perform registration to obtain the initial pseudo labels for sparsely labeled volumes. Subsequently, by introducing unlabeled volumes, we propose a dual-network paradigm named Dense-Sparse Co-training (DeSCO) that exploits dense pseudo labels in early stage and sparse labels in later stage and meanwhile forces consistent output of two networks. Experimental results on three benchmark datasets validated our effectiveness in performance and efficiency in annotation. For example, with only 10 annotated slices, our method reaches a Dice up to 86.93% on KiTS19 dataset.
MT-UDA: Towards Unsupervised Cross-modality Medical Image Segmentation with Limited Source Labels
Mar 23, 2022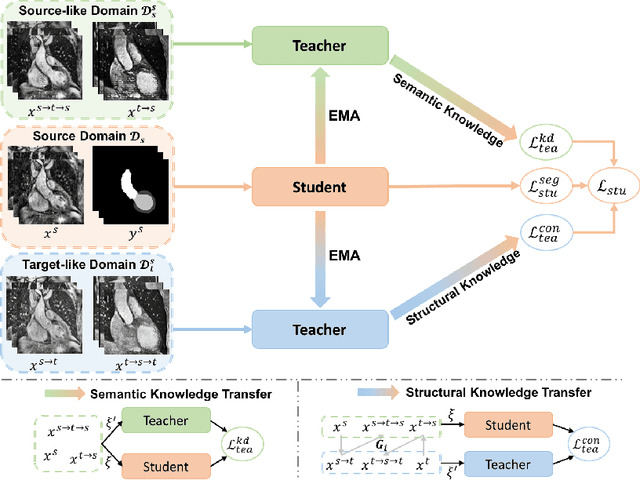
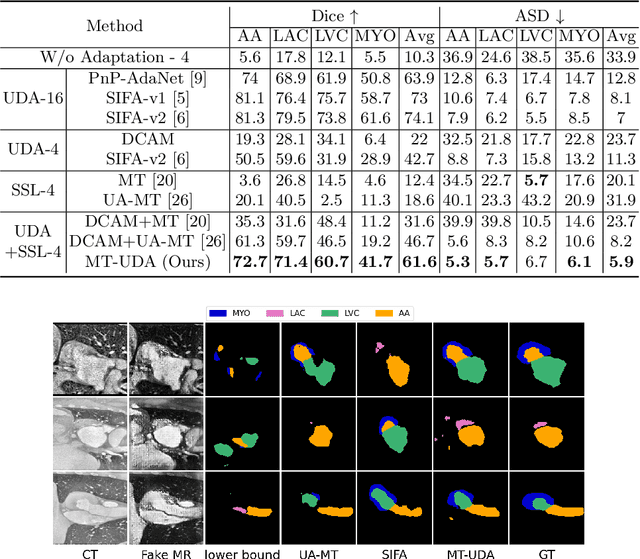
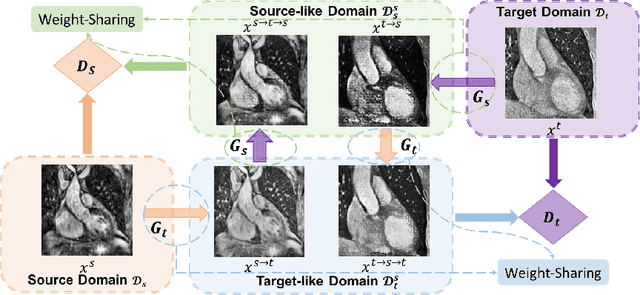
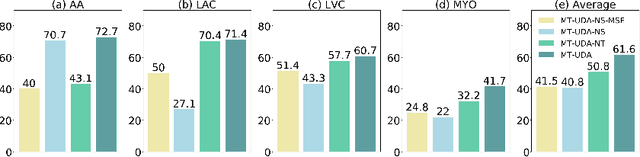
Abstract:The success of deep convolutional neural networks (DCNNs) benefits from high volumes of annotated data. However, annotating medical images is laborious, expensive, and requires human expertise, which induces the label scarcity problem. Especially when encountering the domain shift, the problem becomes more serious. Although deep unsupervised domain adaptation (UDA) can leverage well-established source domain annotations and abundant target domain data to facilitate cross-modality image segmentation and also mitigate the label paucity problem on the target domain, the conventional UDA methods suffer from severe performance degradation when source domain annotations are scarce. In this paper, we explore a challenging UDA setting - limited source domain annotations. We aim to investigate how to efficiently leverage unlabeled data from the source and target domains with limited source annotations for cross-modality image segmentation. To achieve this, we propose a new label-efficient UDA framework, termed MT-UDA, in which the student model trained with limited source labels learns from unlabeled data of both domains by two teacher models respectively in a semi-supervised manner. More specifically, the student model not only distills the intra-domain semantic knowledge by encouraging prediction consistency but also exploits the inter-domain anatomical information by enforcing structural consistency. Consequently, the student model can effectively integrate the underlying knowledge beneath available data resources to mitigate the impact of source label scarcity and yield improved cross-modality segmentation performance. We evaluate our method on MM-WHS 2017 dataset and demonstrate that our approach outperforms the state-of-the-art methods by a large margin under the source-label scarcity scenario.
* Accept by MICCAI 2021, code at: https://github.com/jacobzhaoziyuan/MT-UDA
Hierarchical Consistency Regularized Mean Teacher for Semi-supervised 3D Left Atrium Segmentation
May 21, 2021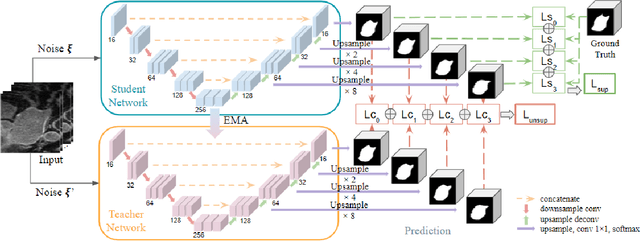
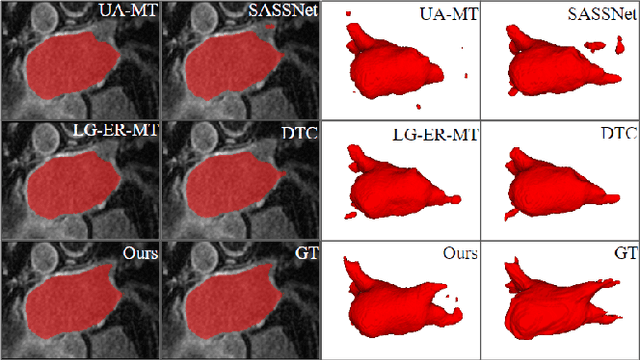
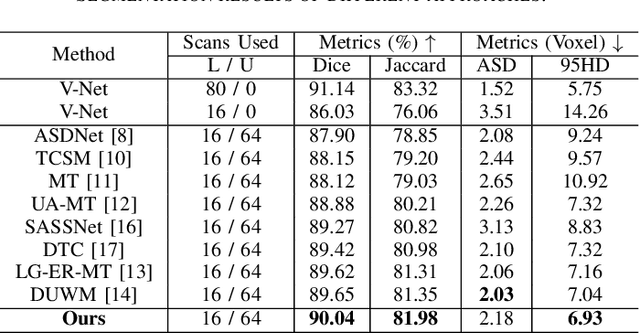
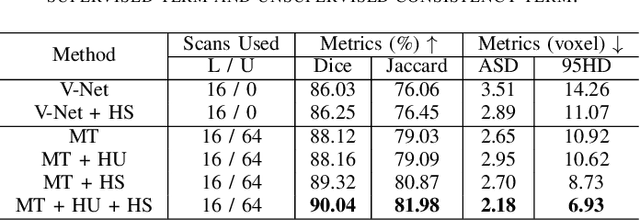
Abstract:Deep learning has achieved promising segmentation performance on 3D left atrium MR images. However, annotations for segmentation tasks are expensive, costly and difficult to obtain. In this paper, we introduce a novel hierarchical consistency regularized mean teacher framework for 3D left atrium segmentation. In each iteration, the student model is optimized by multi-scale deep supervision and hierarchical consistency regularization, concurrently. Extensive experiments have shown that our method achieves competitive performance as compared with full annotation, outperforming other stateof-the-art semi-supervised segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge