Shane Jackson
Synthesis parameter effect detection using quantitative representations and high dimensional distribution distances
Apr 03, 2023Abstract:Detection of effects of the parameters of the synthetic process on the microstructure of materials is an important, yet elusive goal of materials science. We develop a method for detecting effects based on copula theory, high dimensional distribution distances, and permutational statistics to analyze a designed experiment synthesizing plutonium oxide from Pu(III) Oxalate. We detect effects of strike order and oxalic acid feed on the microstructure of the resulting plutonium oxide, which match the literature well. We also detect excess bivariate effects between the pairs of acid concentration, strike order and precipitation temperature.
Do Neural Networks Trained with Topological Features Learn Different Internal Representations?
Nov 14, 2022



Abstract:There is a growing body of work that leverages features extracted via topological data analysis to train machine learning models. While this field, sometimes known as topological machine learning (TML), has seen some notable successes, an understanding of how the process of learning from topological features differs from the process of learning from raw data is still limited. In this work, we begin to address one component of this larger issue by asking whether a model trained with topological features learns internal representations of data that are fundamentally different than those learned by a model trained with the original raw data. To quantify ``different'', we exploit two popular metrics that can be used to measure the similarity of the hidden representations of data within neural networks, neural stitching and centered kernel alignment. From these we draw a range of conclusions about how training with topological features does and does not change the representations that a model learns. Perhaps unsurprisingly, we find that structurally, the hidden representations of models trained and evaluated on topological features differ substantially compared to those trained and evaluated on the corresponding raw data. On the other hand, our experiments show that in some cases, these representations can be reconciled (at least to the degree required to solve the corresponding task) using a simple affine transformation. We conjecture that this means that neural networks trained on raw data may extract some limited topological features in the process of making predictions.
Accelerated Computation of a High Dimensional Kolmogorov-Smirnov Distance
Jun 25, 2021
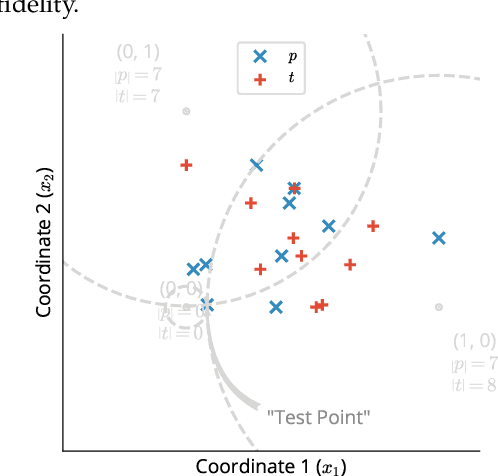


Abstract:Statistical testing is widespread and critical for a variety of scientific disciplines. The advent of machine learning and the increase of computing power has increased the interest in the analysis and statistical testing of multidimensional data. We extend the powerful Kolmogorov-Smirnov two sample test to a high dimensional form in a similar manner to Fasano (Fasano, 1987). We call our result the d-dimensional Kolmogorov-Smirnov test (ddKS) and provide three novel contributions therewith: we develop an analytical equation for the significance of a given ddKS score, we provide an algorithm for computation of ddKS on modern computing hardware that is of constant time complexity for small sample sizes and dimensions, and we provide two approximate calculations of ddKS: one that reduces the time complexity to linear at larger sample sizes, and another that reduces the time complexity to linear with increasing dimension. We perform power analysis of ddKS and its approximations on a corpus of datasets and compare to other common high dimensional two sample tests and distances: Hotelling's T^2 test and Kullback-Leibler divergence. Our ddKS test performs well for all datasets, dimensions, and sizes tested, whereas the other tests and distances fail to reject the null hypothesis on at least one dataset. We therefore conclude that ddKS is a powerful multidimensional two sample test for general use, and can be calculated in a fast and efficient manner using our parallel or approximate methods. Open source implementations of all methods described in this work are located at https://github.com/pnnl/ddks.
Physics-informed Neural-Network Software for Molecular Dynamics Applications
Nov 11, 2020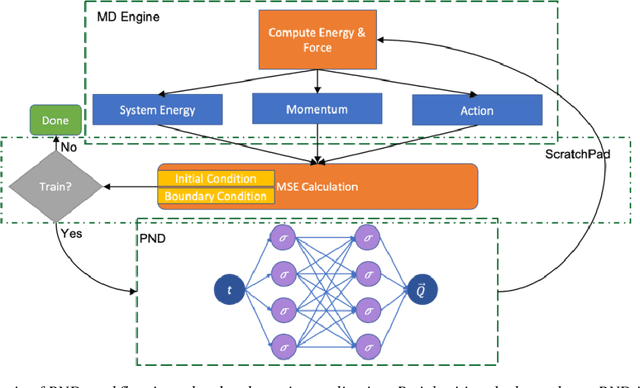

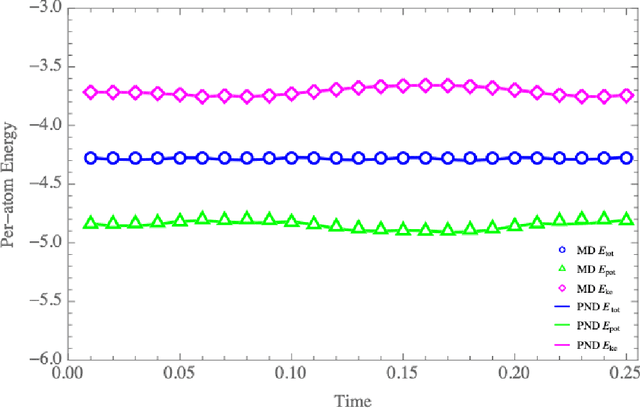
Abstract:We have developed a novel differential equation solver software called PND based on the physics-informed neural network for molecular dynamics simulators. Based on automatic differentiation technique provided by Pytorch, our software allows users to flexibly implement equation of atom motions, initial and boundary conditions, and conservation laws as loss function to train the network. PND comes with a parallel molecular dynamics (MD) engine in order for users to examine and optimize loss function design, and different conservation laws and boundary conditions, and hyperparameters, thereby accelerate the PINN-based development for molecular applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge