Shan Cong
MVKTrans: Multi-View Knowledge Transfer for Robust Multiomics Classification
Nov 13, 2024



Abstract:The distinct characteristics of multiomics data, including complex interactions within and across biological layers and disease heterogeneity (e.g., heterogeneity in etiology and clinical symptoms), drive us to develop novel designs to address unique challenges in multiomics prediction. In this paper, we propose the multi-view knowledge transfer learning (MVKTrans) framework, which transfers intra- and inter-omics knowledge in an adaptive manner by reviewing data heterogeneity and suppressing bias transfer, thereby enhancing classification performance. Specifically, we design a graph contrastive module that is trained on unlabeled data to effectively learn and transfer the underlying intra-omics patterns to the supervised task. This unsupervised pretraining promotes learning general and unbiased representations for each modality, regardless of the downstream tasks. In light of the varying discriminative capacities of modalities across different diseases and/or samples, we introduce an adaptive and bi-directional cross-omics distillation module. This module automatically identifies richer modalities and facilitates dynamic knowledge transfer from more informative to less informative omics, thereby enabling a more robust and generalized integration. Extensive experiments on four real biomedical datasets demonstrate the superior performance and robustness of MVKTrans compared to the state-of-the-art. Code and data are available at https://github.com/Yaolab-fantastic/MVKTrans.
Trustworthy Enhanced Multi-view Multi-modal Alzheimer's Disease Prediction with Brain-wide Imaging Transcriptomics Data
Jun 21, 2024



Abstract:Brain transcriptomics provides insights into the molecular mechanisms by which the brain coordinates its functions and processes. However, existing multimodal methods for predicting Alzheimer's disease (AD) primarily rely on imaging and sometimes genetic data, often neglecting the transcriptomic basis of brain. Furthermore, while striving to integrate complementary information between modalities, most studies overlook the informativeness disparities between modalities. Here, we propose TMM, a trusted multiview multimodal graph attention framework for AD diagnosis, using extensive brain-wide transcriptomics and imaging data. First, we construct view-specific brain regional co-function networks (RRIs) from transcriptomics and multimodal radiomics data to incorporate interaction information from both biomolecular and imaging perspectives. Next, we apply graph attention (GAT) processing to each RRI network to produce graph embeddings and employ cross-modal attention to fuse transcriptomics-derived embedding with each imagingderived embedding. Finally, a novel true-false-harmonized class probability (TFCP) strategy is designed to assess and adaptively adjust the prediction confidence of each modality for AD diagnosis. We evaluate TMM using the AHBA database with brain-wide transcriptomics data and the ADNI database with three imaging modalities (AV45-PET, FDG-PET, and VBM-MRI). The results demonstrate the superiority of our method in identifying AD, EMCI, and LMCI compared to state-of-the-arts. Code and data are available at https://github.com/Yaolab-fantastic/TMM.
Image Registration and Predictive Modeling: Learning the Metric on the Space of Diffeomorphisms
Aug 10, 2018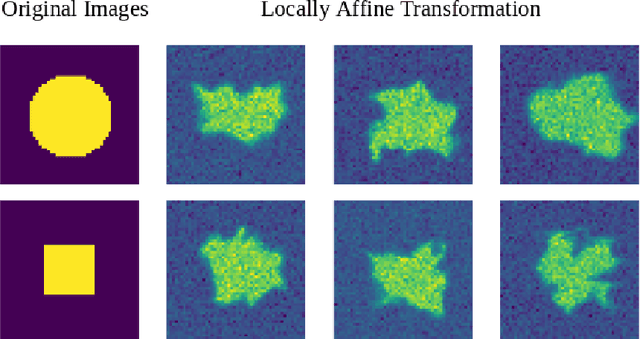

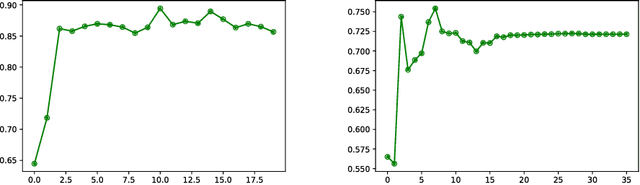
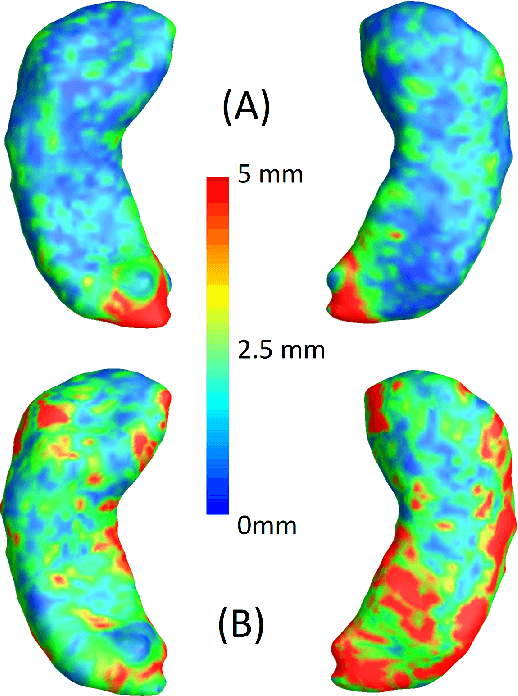
Abstract:We present a method for metric optimization in the Large Deformation Diffeomorphic Metric Mapping (LDDMM) framework, by treating the induced Riemannian metric on the space of diffeomorphisms as a kernel in a machine learning context. For simplicity, we choose the kernel Fischer Linear Discriminant Analysis (KLDA) as the framework. Optimizing the kernel parameters in an Expectation-Maximization framework, we define model fidelity via the hinge loss of the decision function. The resulting algorithm optimizes the parameters of the LDDMM norm-inducing differential operator as a solution to a group-wise registration and classification problem. In practice, this may lead to a biology-aware registration, focusing its attention on the predictive task at hand such as identifying the effects of disease. We first tested our algorithm on a synthetic dataset, showing that our parameter selection improves registration quality and classification accuracy. We then tested the algorithm on 3D subcortical shapes from the Schizophrenia cohort Schizconnect. Our Schizpohrenia-Control predictive model showed significant improvement in ROC AUC compared to baseline parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge