Seungbeom Lee
Plane Geometry Problem Solving with Multi-modal Reasoning: A Survey
May 20, 2025Abstract:Plane geometry problem solving (PGPS) has recently gained significant attention as a benchmark to assess the multi-modal reasoning capabilities of large vision-language models. Despite the growing interest in PGPS, the research community still lacks a comprehensive overview that systematically synthesizes recent work in PGPS. To fill this gap, we present a survey of existing PGPS studies. We first categorize PGPS methods into an encoder-decoder framework and summarize the corresponding output formats used by their encoders and decoders. Subsequently, we classify and analyze these encoders and decoders according to their architectural designs. Finally, we outline major challenges and promising directions for future research. In particular, we discuss the hallucination issues arising during the encoding phase within encoder-decoder architectures, as well as the problem of data leakage in current PGPS benchmarks.
GeoDANO: Geometric VLM with Domain Agnostic Vision Encoder
Feb 17, 2025Abstract:We introduce GeoDANO, a geometric vision-language model (VLM) with a domain-agnostic vision encoder, for solving plane geometry problems. Although VLMs have been employed for solving geometry problems, their ability to recognize geometric features remains insufficiently analyzed. To address this gap, we propose a benchmark that evaluates the recognition of visual geometric features, including primitives such as dots and lines, and relations such as orthogonality. Our preliminary study shows that vision encoders often used in general-purpose VLMs, e.g., OpenCLIP, fail to detect these features and struggle to generalize across domains. We develop GeoCLIP, a CLIP based model trained on synthetic geometric diagram-caption pairs to overcome the limitation. Benchmark results show that GeoCLIP outperforms existing vision encoders in recognizing geometric features. We then propose our VLM, GeoDANO, which augments GeoCLIP with a domain adaptation strategy for unseen diagram styles. GeoDANO outperforms specialized methods for plane geometry problems and GPT-4o on MathVerse.
EPIC: Graph Augmentation with Edit Path Interpolation via Learnable Cost
Jun 02, 2023Abstract:Graph-based models have become increasingly important in various domains, but the limited size and diversity of existing graph datasets often limit their performance. To address this issue, we propose EPIC (Edit Path Interpolation via learnable Cost), a novel interpolation-based method for augmenting graph datasets. Our approach leverages graph edit distance to generate new graphs that are similar to the original ones but exhibit some variation in their structures. To achieve this, we learn the graph edit distance through a comparison of labeled graphs and utilize this knowledge to create graph edit paths between pairs of original graphs. With randomly sampled graphs from a graph edit path, we enrich the training set to enhance the generalization capability of classification models. We demonstrate the effectiveness of our approach on several benchmark datasets and show that it outperforms existing augmentation methods in graph classification tasks.
Substructure-Atom Cross Attention for Molecular Representation Learning
Oct 15, 2022


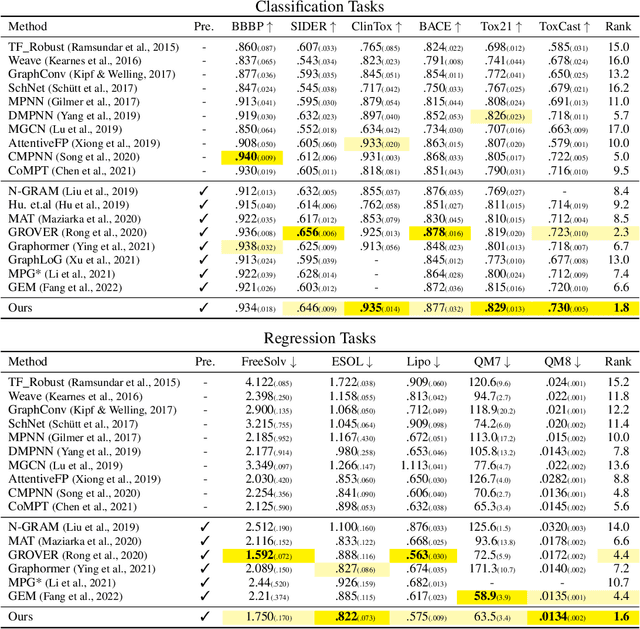
Abstract:Designing a neural network architecture for molecular representation is crucial for AI-driven drug discovery and molecule design. In this work, we propose a new framework for molecular representation learning. Our contribution is threefold: (a) demonstrating the usefulness of incorporating substructures to node-wise features from molecules, (b) designing two branch networks consisting of a transformer and a graph neural network so that the networks fused with asymmetric attention, and (c) not requiring heuristic features and computationally-expensive information from molecules. Using 1.8 million molecules collected from ChEMBL and PubChem database, we pretrain our network to learn a general representation of molecules with minimal supervision. The experimental results show that our pretrained network achieves competitive performance on 11 downstream tasks for molecular property prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge