Sascha Jecklin
IXGS-Intraoperative 3D Reconstruction from Sparse, Arbitrarily Posed Real X-rays
Apr 20, 2025Abstract:Spine surgery is a high-risk intervention demanding precise execution, often supported by image-based navigation systems. Recently, supervised learning approaches have gained attention for reconstructing 3D spinal anatomy from sparse fluoroscopic data, significantly reducing reliance on radiation-intensive 3D imaging systems. However, these methods typically require large amounts of annotated training data and may struggle to generalize across varying patient anatomies or imaging conditions. Instance-learning approaches like Gaussian splatting could offer an alternative by avoiding extensive annotation requirements. While Gaussian splatting has shown promise for novel view synthesis, its application to sparse, arbitrarily posed real intraoperative X-rays has remained largely unexplored. This work addresses this limitation by extending the $R^2$-Gaussian splatting framework to reconstruct anatomically consistent 3D volumes under these challenging conditions. We introduce an anatomy-guided radiographic standardization step using style transfer, improving visual consistency across views, and enhancing reconstruction quality. Notably, our framework requires no pretraining, making it inherently adaptable to new patients and anatomies. We evaluated our approach using an ex-vivo dataset. Expert surgical evaluation confirmed the clinical utility of the 3D reconstructions for navigation, especially when using 20 to 30 views, and highlighted the standardization's benefit for anatomical clarity. Benchmarking via quantitative 2D metrics (PSNR/SSIM) confirmed performance trade-offs compared to idealized settings, but also validated the improvement gained from standardization over raw inputs. This work demonstrates the feasibility of instance-based volumetric reconstruction from arbitrary sparse-view X-rays, advancing intraoperative 3D imaging for surgical navigation.
ArthroPhase: A Novel Dataset and Method for Phase Recognition in Arthroscopic Video
Feb 11, 2025



Abstract:This study aims to advance surgical phase recognition in arthroscopic procedures, specifically Anterior Cruciate Ligament (ACL) reconstruction, by introducing the first arthroscopy dataset and developing a novel transformer-based model. We aim to establish a benchmark for arthroscopic surgical phase recognition by leveraging spatio-temporal features to address the specific challenges of arthroscopic videos including limited field of view, occlusions, and visual distortions. We developed the ACL27 dataset, comprising 27 videos of ACL surgeries, each labeled with surgical phases. Our model employs a transformer-based architecture, utilizing temporal-aware frame-wise feature extraction through a ResNet-50 and transformer layers. This approach integrates spatio-temporal features and introduces a Surgical Progress Index (SPI) to quantify surgery progression. The model's performance was evaluated using accuracy, precision, recall, and Jaccard Index on the ACL27 and Cholec80 datasets. The proposed model achieved an overall accuracy of 72.91% on the ACL27 dataset. On the Cholec80 dataset, the model achieved a comparable performance with the state-of-the-art methods with an accuracy of 92.4%. The SPI demonstrated an output error of 10.6% and 9.86% on ACL27 and Cholec80 datasets respectively, indicating reliable surgery progression estimation. This study introduces a significant advancement in surgical phase recognition for arthroscopy, providing a comprehensive dataset and a robust transformer-based model. The results validate the model's effectiveness and generalizability, highlighting its potential to improve surgical training, real-time assistance, and operational efficiency in orthopedic surgery. The publicly available dataset and code will facilitate future research and development in this critical field.
SurgPointTransformer: Vertebrae Shape Completion with RGB-D Data
Oct 02, 2024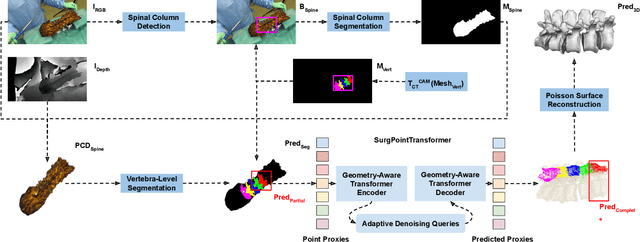
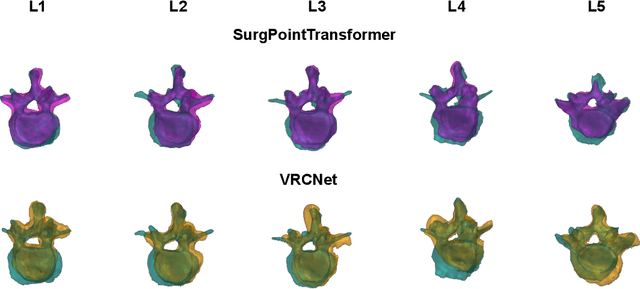
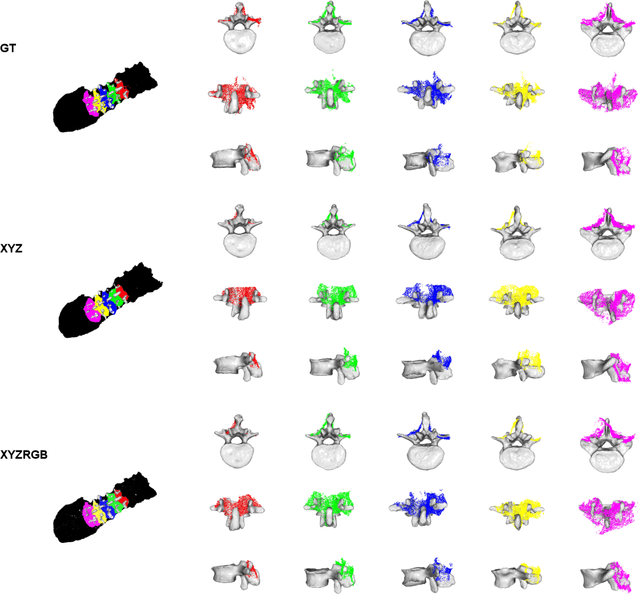
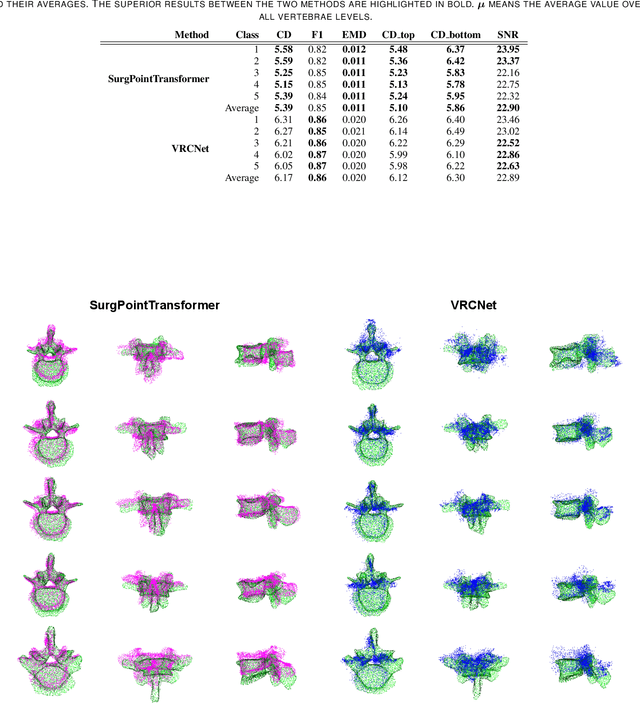
Abstract:State-of-the-art computer- and robot-assisted surgery systems heavily depend on intraoperative imaging technologies such as CT and fluoroscopy to generate detailed 3D visualization of the patient's anatomy. While imaging techniques are highly accurate, they are based on ionizing radiation and expose patients and clinicians. This study introduces an alternative, radiation-free approach for reconstructing the 3D spine anatomy using RGB-D data. Drawing inspiration from the 3D "mental map" that surgeons form during surgeries, we introduce SurgPointTransformer, a shape completion approach for surgical applications that can accurately reconstruct the unexposed spine regions from sparse observations of the exposed surface. Our method involves two main steps: segmentation and shape completion. The segmentation step includes spinal column localization and segmentation, followed by vertebra-wise segmentation. The segmented vertebra point clouds are then subjected to SurgPointTransformer, which leverages an attention mechanism to learn patterns between visible surface features and the underlying anatomy. For evaluation, we utilize an ex-vivo dataset of nine specimens. Their CT data is used to establish ground truth data that were used to compare to the outputs of our methods. Our method significantly outperforms the state-of-the-art baselines, achieving an average Chamfer Distance of 5.39, an F-Score of 0.85, an Earth Mover's Distance of 0.011, and a Signal-to-Noise Ratio of 22.90 dB. This study demonstrates the potential of our reconstruction method for 3D vertebral shape completion. It enables 3D reconstruction of the entire lumbar spine and surgical guidance without ionizing radiation or invasive imaging. Our work contributes to computer-aided and robot-assisted surgery, advancing the perception and intelligence of these systems.
Domain adaptation strategies for 3D reconstruction of the lumbar spine using real fluoroscopy data
Jan 29, 2024

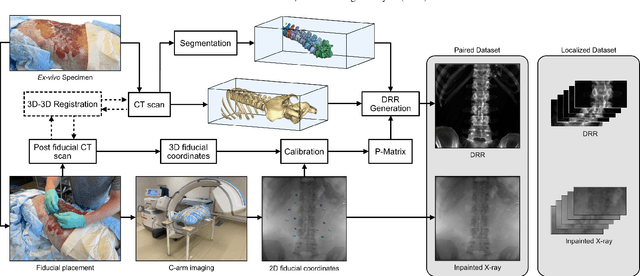

Abstract:This study tackles key obstacles in adopting surgical navigation in orthopedic surgeries, including time, cost, radiation, and workflow integration challenges. Recently, our work X23D showed an approach for generating 3D anatomical models of the spine from only a few intraoperative fluoroscopic images. This negates the need for conventional registration-based surgical navigation by creating a direct intraoperative 3D reconstruction of the anatomy. Despite these strides, the practical application of X23D has been limited by a domain gap between synthetic training data and real intraoperative images. In response, we devised a novel data collection protocol for a paired dataset consisting of synthetic and real fluoroscopic images from the same perspectives. Utilizing this dataset, we refined our deep learning model via transfer learning, effectively bridging the domain gap between synthetic and real X-ray data. A novel style transfer mechanism also allows us to convert real X-rays to mirror the synthetic domain, enabling our in-silico-trained X23D model to achieve high accuracy in real-world settings. Our results demonstrated that the refined model can rapidly generate accurate 3D reconstructions of the entire lumbar spine from as few as three intraoperative fluoroscopic shots. It achieved an 84% F1 score, matching the accuracy of our previous synthetic data-based research. Additionally, with a computational time of only 81.1 ms, our approach provides real-time capabilities essential for surgery integration. Through examining ideal imaging setups and view angle dependencies, we've further confirmed our system's practicality and dependability in clinical settings. Our research marks a significant step forward in intraoperative 3D reconstruction, offering enhancements to surgical planning, navigation, and robotics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge