Aidana Massalimova
NeuralBoneReg: A Novel Self-Supervised Method for Robust and Accurate Multi-Modal Bone Surface Registration
Nov 18, 2025Abstract:In computer- and robot-assisted orthopedic surgery (CAOS), patient-specific surgical plans derived from preoperative imaging define target locations and implant trajectories. During surgery, these plans must be accurately transferred, relying on precise cross-registration between preoperative and intraoperative data. However, substantial modality heterogeneity across imaging modalities makes this registration challenging and error-prone. Robust, automatic, and modality-agnostic bone surface registration is therefore clinically important. We propose NeuralBoneReg, a self-supervised, surface-based framework that registers bone surfaces using 3D point clouds as a modality-agnostic representation. NeuralBoneReg includes two modules: an implicit neural unsigned distance field (UDF) that learns the preoperative bone model, and an MLP-based registration module that performs global initialization and local refinement by generating transformation hypotheses to align the intraoperative point cloud with the neural UDF. Unlike SOTA supervised methods, NeuralBoneReg operates in a self-supervised manner, without requiring inter-subject training data. We evaluated NeuralBoneReg against baseline methods on two publicly available multi-modal datasets: a CT-ultrasound dataset of the fibula and tibia (UltraBones100k) and a CT-RGB-D dataset of spinal vertebrae (SpineDepth). The evaluation also includes a newly introduced CT--ultrasound dataset of cadaveric subjects containing femur and pelvis (UltraBones-Hip), which will be made publicly available. NeuralBoneReg matches or surpasses existing methods across all datasets, achieving mean RRE/RTE of 1.68°/1.86 mm on UltraBones100k, 1.88°/1.89 mm on UltraBones-Hip, and 3.79°/2.45 mm on SpineDepth. These results demonstrate strong generalizability across anatomies and modalities, providing robust and accurate cross-modal alignment for CAOS.
IXGS-Intraoperative 3D Reconstruction from Sparse, Arbitrarily Posed Real X-rays
Apr 20, 2025Abstract:Spine surgery is a high-risk intervention demanding precise execution, often supported by image-based navigation systems. Recently, supervised learning approaches have gained attention for reconstructing 3D spinal anatomy from sparse fluoroscopic data, significantly reducing reliance on radiation-intensive 3D imaging systems. However, these methods typically require large amounts of annotated training data and may struggle to generalize across varying patient anatomies or imaging conditions. Instance-learning approaches like Gaussian splatting could offer an alternative by avoiding extensive annotation requirements. While Gaussian splatting has shown promise for novel view synthesis, its application to sparse, arbitrarily posed real intraoperative X-rays has remained largely unexplored. This work addresses this limitation by extending the $R^2$-Gaussian splatting framework to reconstruct anatomically consistent 3D volumes under these challenging conditions. We introduce an anatomy-guided radiographic standardization step using style transfer, improving visual consistency across views, and enhancing reconstruction quality. Notably, our framework requires no pretraining, making it inherently adaptable to new patients and anatomies. We evaluated our approach using an ex-vivo dataset. Expert surgical evaluation confirmed the clinical utility of the 3D reconstructions for navigation, especially when using 20 to 30 views, and highlighted the standardization's benefit for anatomical clarity. Benchmarking via quantitative 2D metrics (PSNR/SSIM) confirmed performance trade-offs compared to idealized settings, but also validated the improvement gained from standardization over raw inputs. This work demonstrates the feasibility of instance-based volumetric reconstruction from arbitrary sparse-view X-rays, advancing intraoperative 3D imaging for surgical navigation.
SurgPointTransformer: Vertebrae Shape Completion with RGB-D Data
Oct 02, 2024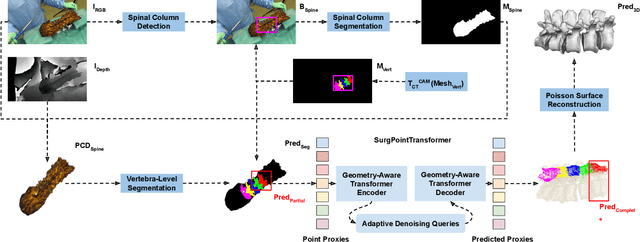
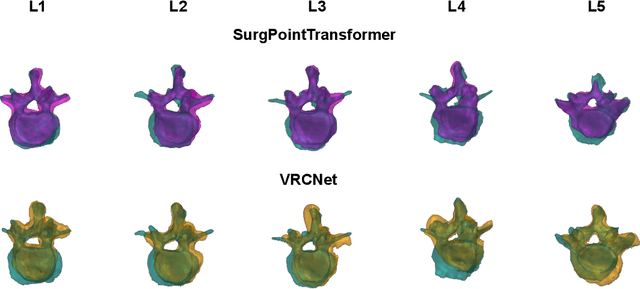
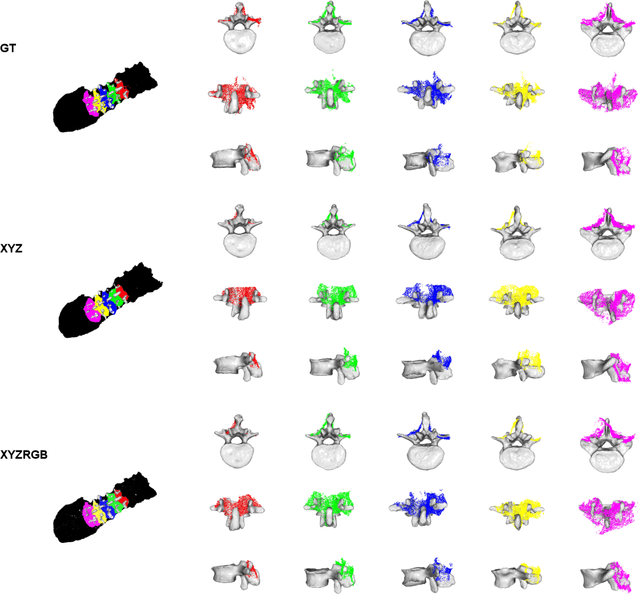
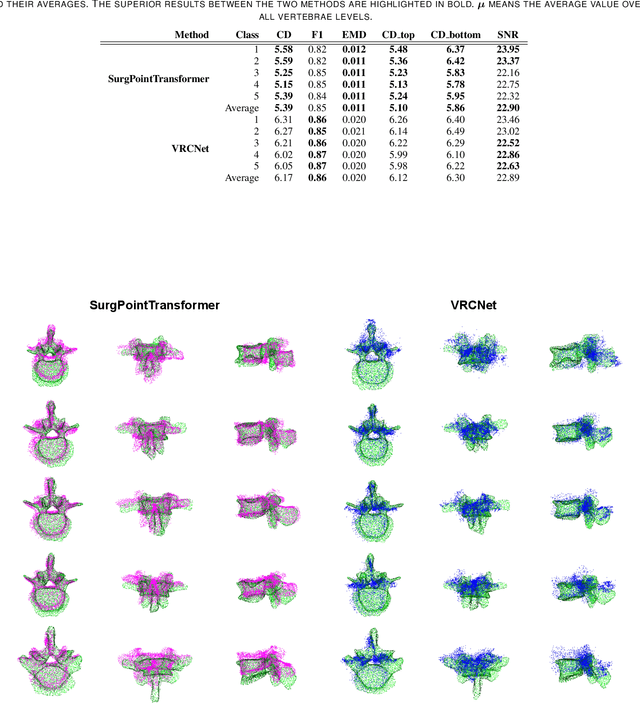
Abstract:State-of-the-art computer- and robot-assisted surgery systems heavily depend on intraoperative imaging technologies such as CT and fluoroscopy to generate detailed 3D visualization of the patient's anatomy. While imaging techniques are highly accurate, they are based on ionizing radiation and expose patients and clinicians. This study introduces an alternative, radiation-free approach for reconstructing the 3D spine anatomy using RGB-D data. Drawing inspiration from the 3D "mental map" that surgeons form during surgeries, we introduce SurgPointTransformer, a shape completion approach for surgical applications that can accurately reconstruct the unexposed spine regions from sparse observations of the exposed surface. Our method involves two main steps: segmentation and shape completion. The segmentation step includes spinal column localization and segmentation, followed by vertebra-wise segmentation. The segmented vertebra point clouds are then subjected to SurgPointTransformer, which leverages an attention mechanism to learn patterns between visible surface features and the underlying anatomy. For evaluation, we utilize an ex-vivo dataset of nine specimens. Their CT data is used to establish ground truth data that were used to compare to the outputs of our methods. Our method significantly outperforms the state-of-the-art baselines, achieving an average Chamfer Distance of 5.39, an F-Score of 0.85, an Earth Mover's Distance of 0.011, and a Signal-to-Noise Ratio of 22.90 dB. This study demonstrates the potential of our reconstruction method for 3D vertebral shape completion. It enables 3D reconstruction of the entire lumbar spine and surgical guidance without ionizing radiation or invasive imaging. Our work contributes to computer-aided and robot-assisted surgery, advancing the perception and intelligence of these systems.
Automatic breach detection during spine pedicle drilling based on vibroacoustic sensing
Mar 27, 2023



Abstract:Pedicle drilling is a complex and critical spinal surgery task. Detecting breach or penetration of the surgical tool to the cortical wall during pilot-hole drilling is essential to avoid damage to vital anatomical structures adjacent to the pedicle, such as the spinal cord, blood vessels, and nerves. Currently, the guidance of pedicle drilling is done using image-guided methods that are radiation intensive and limited to the preoperative information. This work proposes a new radiation-free breach detection algorithm leveraging a non-visual sensor setup in combination with deep learning approach. Multiple vibroacoustic sensors, such as a contact microphone, a free-field microphone, a tri-axial accelerometer, a uni-axial accelerometer, and an optical tracking system were integrated into the setup. Data were collected on four cadaveric human spines, ranging from L5 to T10. An experienced spine surgeon drilled the pedicles relying on optical navigation. A new automatic labeling method based on the tracking data was introduced. Labeled data was subsequently fed to the network in mel-spectrograms, classifying the data into breach and non-breach. Different sensor types, sensor positioning, and their combinations were evaluated. The best results in breach recall for individual sensors could be achieved using contact microphones attached to the dorsal skin (85.8\%) and uni-axial accelerometers clamped to the spinous process of the drilled vertebra (81.0\%). The best-performing data fusion model combined the latter two sensors with a breach recall of 98\%. The proposed method shows the great potential of non-visual sensor fusion for avoiding screw misplacement and accidental bone breaches during pedicle drilling and could be extended to further surgical applications.
Input Agnostic Deep Learning for Alzheimer's Disease Classification Using Multimodal MRI Images
Jul 19, 2021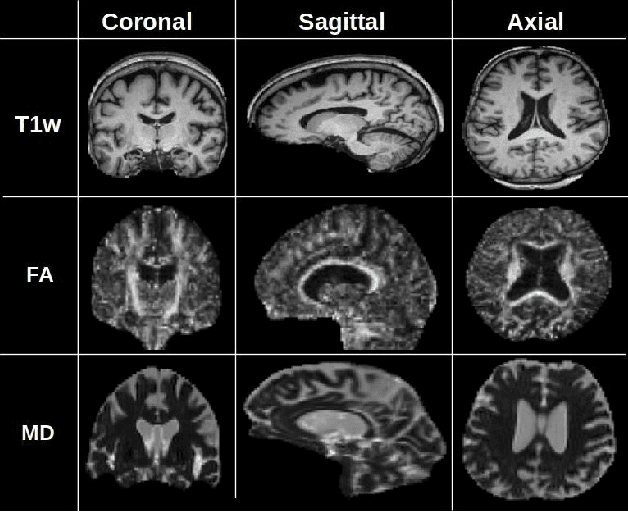
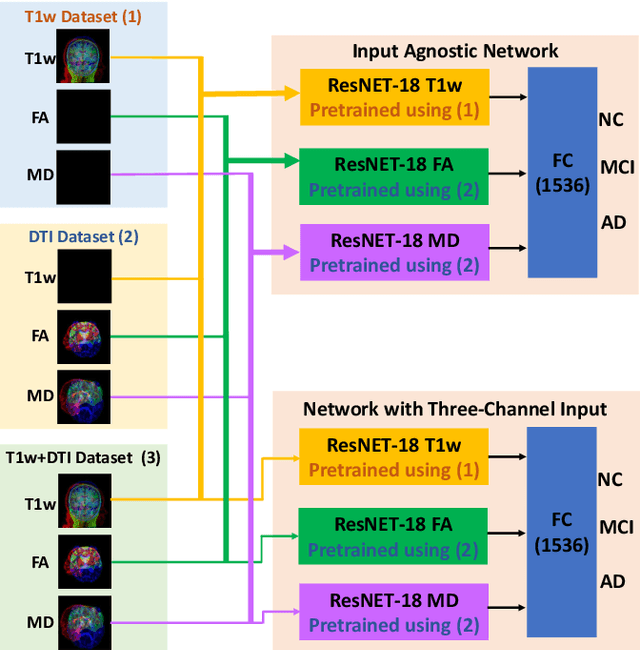


Abstract:Alzheimer's disease (AD) is a progressive brain disorder that causes memory and functional impairments. The advances in machine learning and publicly available medical datasets initiated multiple studies in AD diagnosis. In this work, we utilize a multi-modal deep learning approach in classifying normal cognition, mild cognitive impairment and AD classes on the basis of structural MRI and diffusion tensor imaging (DTI) scans from the OASIS-3 dataset. In addition to a conventional multi-modal network, we also present an input agnostic architecture that allows diagnosis with either sMRI or DTI scan, which distinguishes our method from previous multi-modal machine learning-based methods. The results show that the input agnostic model achieves 0.96 accuracy when both structural MRI and DTI scans are provided as inputs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge