Salman Seyedi
Department of Biomedical Informatics, Emory University, Atlanta, USA
Detection of Chagas Disease from the ECG: The George B. Moody PhysioNet Challenge 2025
Oct 02, 2025Abstract:Objective: Chagas disease is a parasitic infection that is endemic to South America, Central America, and, more recently, the U.S., primarily transmitted by insects. Chronic Chagas disease can cause cardiovascular diseases and digestive problems. Serological testing capacities for Chagas disease are limited, but Chagas cardiomyopathy often manifests in ECGs, providing an opportunity to prioritize patients for testing and treatment. Approach: The George B. Moody PhysioNet Challenge 2025 invites teams to develop algorithmic approaches for identifying Chagas disease from electrocardiograms (ECGs). Main results: This Challenge provides multiple innovations. First, we leveraged several datasets with labels from patient reports and serological testing, provided a large dataset with weak labels and smaller datasets with strong labels. Second, we augmented the data to support model robustness and generalizability to unseen data sources. Third, we applied an evaluation metric that captured the local serological testing capacity for Chagas disease to frame the machine learning problem as a triage task. Significance: Over 630 participants from 111 teams submitted over 1300 entries during the Challenge, representing diverse approaches from academia and industry worldwide.
Detecting Cognitive Impairment and Psychological Well-being among Older Adults Using Facial, Acoustic, Linguistic, and Cardiovascular Patterns Derived from Remote Conversations
Dec 23, 2024



Abstract:The aging society urgently requires scalable methods to monitor cognitive decline and identify social and psychological factors indicative of dementia risk in older adults. Our machine learning (ML) models captured facial, acoustic, linguistic, and cardiovascular features from 39 individuals with normal cognition or Mild Cognitive Impairment derived from remote video conversations and classified cognitive status, social isolation, neuroticism, and psychological well-being. Our model could distinguish Clinical Dementia Rating Scale (CDR) of 0.5 (vs. 0) with 0.78 area under the receiver operating characteristic curve (AUC), social isolation with 0.75 AUC, neuroticism with 0.71 AUC, and negative affect scales with 0.79 AUC. Recent advances in machine learning offer new opportunities to remotely detect cognitive impairment and assess associated factors, such as neuroticism and psychological well-being. Our experiment showed that speech and language patterns were more useful for quantifying cognitive impairment, whereas facial expression and cardiovascular patterns using photoplethysmography (PPG) were more useful for quantifying personality and psychological well-being.
ECG-Image-Database: A Dataset of ECG Images with Real-World Imaging and Scanning Artifacts; A Foundation for Computerized ECG Image Digitization and Analysis
Sep 25, 2024

Abstract:We introduce the ECG-Image-Database, a large and diverse collection of electrocardiogram (ECG) images generated from ECG time-series data, with real-world scanning, imaging, and physical artifacts. We used ECG-Image-Kit, an open-source Python toolkit, to generate realistic images of 12-lead ECG printouts from raw ECG time-series. The images include realistic distortions such as noise, wrinkles, stains, and perspective shifts, generated both digitally and physically. The toolkit was applied to 977 12-lead ECG records from the PTB-XL database and 1,000 from Emory Healthcare to create high-fidelity synthetic ECG images. These unique images were subjected to both programmatic distortions using ECG-Image-Kit and physical effects like soaking, staining, and mold growth, followed by scanning and photography under various lighting conditions to create real-world artifacts. The resulting dataset includes 35,595 software-labeled ECG images with a wide range of imaging artifacts and distortions. The dataset provides ground truth time-series data alongside the images, offering a reference for developing machine and deep learning models for ECG digitization and classification. The images vary in quality, from clear scans of clean papers to noisy photographs of degraded papers, enabling the development of more generalizable digitization algorithms. ECG-Image-Database addresses a critical need for digitizing paper-based and non-digital ECGs for computerized analysis, providing a foundation for developing robust machine and deep learning models capable of converting ECG images into time-series. The dataset aims to serve as a reference for ECG digitization and computerized annotation efforts. ECG-Image-Database was used in the PhysioNet Challenge 2024 on ECG image digitization and classification.
Privacy-Preserving Eye-tracking Using Deep Learning
Jun 22, 2021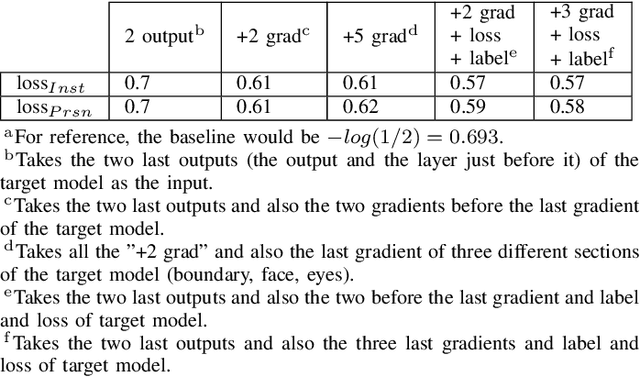
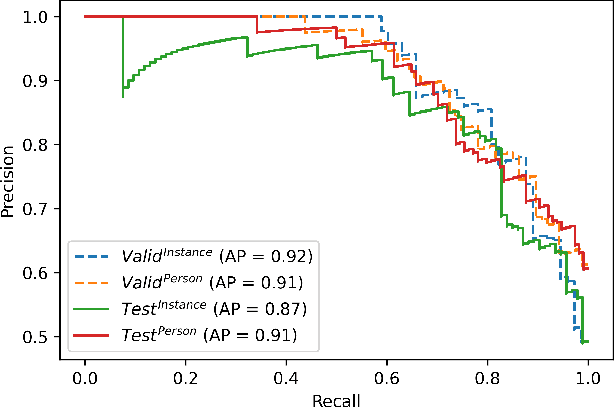
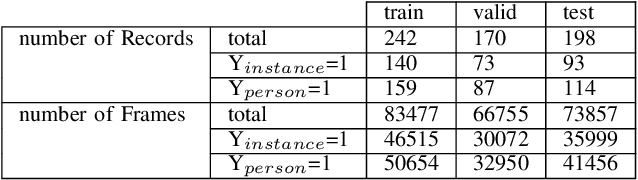
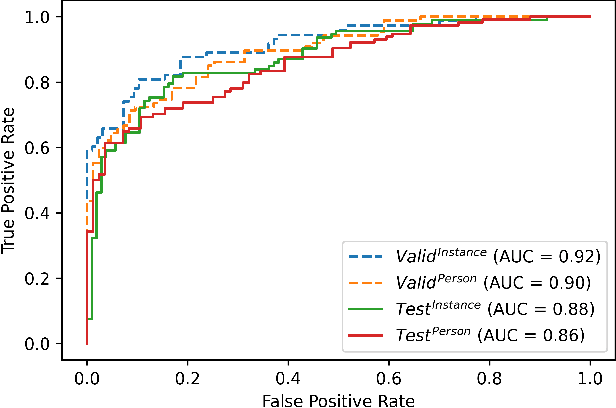
Abstract:The expanding usage of complex machine learning methods like deep learning has led to an explosion in human activity recognition, particularly applied to health. In particular, as part of a larger body sensor network system, face and full-body analysis is becoming increasingly common for evaluating health status. However, complex models which handle private and sometimes protected data, raise concerns about the potential leak of identifiable data. In this work, we focus on the case of a deep network model trained on images of individual faces. Full-face video recordings taken from 493 individuals undergoing an eye-tracking based evaluation of neurological function were used. Outputs, gradients, intermediate layer outputs, loss, and labels were used as inputs for a deep network with an added support vector machine emission layer to recognize membership in the training data. The inference attack method and associated mathematical analysis indicate that there is a low likelihood of unintended memorization of facial features in the deep learning model. In this study, it is showed that the named model preserves the integrity of training data with reasonable confidence. The same process can be implemented in similar conditions for different models.
An Analysis Of Protected Health Information Leakage In Deep-Learning Based De-Identification Algorithms
Jan 28, 2021
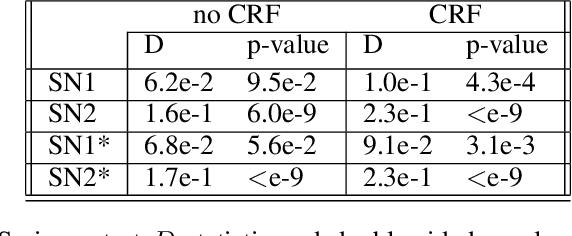
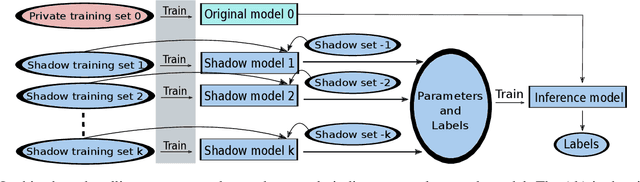
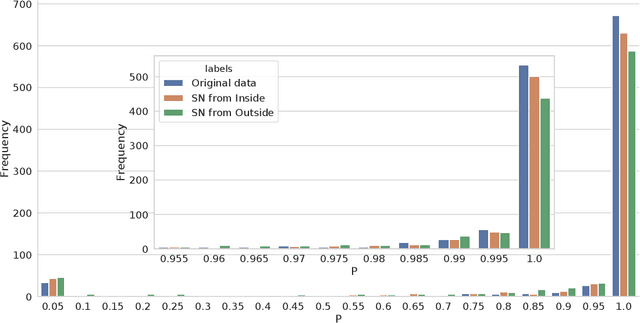
Abstract:The increasing complexity of algorithms for analyzing medical data, including de-identification tasks, raises the possibility that complex algorithms are learning not just the general representation of the problem, but specifics of given individuals within the data. Modern legal frameworks specifically prohibit the intentional or accidental distribution of patient data, but have not addressed this potential avenue for leakage of such protected health information. Modern deep learning algorithms have the highest potential of such leakage due to complexity of the models. Recent research in the field has highlighted such issues in non-medical data, but all analysis is likely to be data and algorithm specific. We, therefore, chose to analyze a state-of-the-art free-text de-identification algorithm based on LSTM (Long Short-Term Memory) and its potential in encoding any individual in the training set. Using the i2b2 Challenge Data, we trained, then analyzed the model to assess whether the output of the LSTM, before the compression layer of the classifier, could be used to estimate the membership of the training data. Furthermore, we used different attacks including membership inference attack method to attack the model. Results indicate that the attacks could not identify whether members of the training data were distinguishable from non-members based on the model output. This indicates that the model does not provide any strong evidence into the identification of the individuals in the training data set and there is not yet empirical evidence it is unsafe to distribute the model for general use.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge