Said Boumaraf
Deep Distance Map Regression Network with Shape-aware Loss for Imbalanced Medical Image Segmentation
Jan 15, 2025Abstract:Small object segmentation, like tumor segmentation, is a difficult and critical task in the field of medical image analysis. Although deep learning based methods have achieved promising performance, they are restricted to the use of binary segmentation mask. Inspired by the rigorous mapping between binary segmentation mask and distance map, we adopt distance map as a novel ground truth and employ a network to fulfill the computation of distance map. Specially, we propose a new segmentation framework that incorporates the existing binary segmentation network and a light weight regression network (dubbed as LR-Net). Thus, the LR-Net can convert the distance map computation into a regression task and leverage the rich information of distance maps. Additionally, we derive a shape-aware loss by employing distance maps as penalty map to infer the complete shape of an object. We evaluated our approach on MICCAI 2017 Liver Tumor Segmentation (LiTS) Challenge dataset and a clinical dataset. Experimental results show that our approach outperforms the classification-based methods as well as other existing state-of-the-arts.
* Conference
BENet: A Cross-domain Robust Network for Detecting Face Forgeries via Bias Expansion and Latent-space Attention
Dec 10, 2024Abstract:In response to the growing threat of deepfake technology, we introduce BENet, a Cross-Domain Robust Bias Expansion Network. BENet enhances the detection of fake faces by addressing limitations in current detectors related to variations across different types of fake face generation techniques, where ``cross-domain" refers to the diverse range of these deepfakes, each considered a separate domain. BENet's core feature is a bias expansion module based on autoencoders. This module maintains genuine facial features while enhancing differences in fake reconstructions, creating a reliable bias for detecting fake faces across various deepfake domains. We also introduce a Latent-Space Attention (LSA) module to capture inconsistencies related to fake faces at different scales, ensuring robust defense against advanced deepfake techniques. The enriched LSA feature maps are multiplied with the expanded bias to create a versatile feature space optimized for subtle forgeries detection. To improve its ability to detect fake faces from unknown sources, BENet integrates a cross-domain detector module that enhances recognition accuracy by verifying the facial domain during inference. We train our network end-to-end with a novel bias expansion loss, adopted for the first time, in face forgery detection. Extensive experiments covering both intra and cross-dataset demonstrate BENet's superiority over current state-of-the-art solutions.
Reconstructing Deep Neural Networks: Unleashing the Optimization Potential of Natural Gradient Descent
Dec 10, 2024



Abstract:Natural gradient descent (NGD) is a powerful optimization technique for machine learning, but the computational complexity of the inverse Fisher information matrix limits its application in training deep neural networks. To overcome this challenge, we propose a novel optimization method for training deep neural networks called structured natural gradient descent (SNGD). Theoretically, we demonstrate that optimizing the original network using NGD is equivalent to using fast gradient descent (GD) to optimize the reconstructed network with a structural transformation of the parameter matrix. Thereby, we decompose the calculation of the global Fisher information matrix into the efficient computation of local Fisher matrices via constructing local Fisher layers in the reconstructed network to speed up the training. Experimental results on various deep networks and datasets demonstrate that SNGD achieves faster convergence speed than NGD while retaining comparable solutions. Furthermore, our method outperforms traditional GDs in terms of efficiency and effectiveness. Thus, our proposed method has the potential to significantly improve the scalability and efficiency of NGD in deep learning applications. Our source code is available at https://github.com/Chaochao-Lin/SNGD.
Dynamic Multi-Domain Knowledge Networks for Chest X-ray Report Generation
Oct 08, 2023Abstract:The automated generation of radiology diagnostic reports helps radiologists make timely and accurate diagnostic decisions while also enhancing clinical diagnostic efficiency. However, the significant imbalance in the distribution of data between normal and abnormal samples (including visual and textual biases) poses significant challenges for a data-driven task like automatically generating diagnostic radiology reports. Therefore, we propose a Dynamic Multi-Domain Knowledge(DMDK) network for radiology diagnostic report generation. The DMDK network consists of four modules: Chest Feature Extractor(CFE), Dynamic Knowledge Extractor(DKE), Specific Knowledge Extractor(SKE), and Multi-knowledge Integrator(MKI) module. Specifically, the CFE module is primarily responsible for extracting the unprocessed visual medical features of the images. The DKE module is responsible for extracting dynamic disease topic labels from the retrieved radiology diagnostic reports. We then fuse the dynamic disease topic labels with the original visual features of the images to highlight the abnormal regions in the original visual features to alleviate the visual data bias problem. The SKE module expands upon the conventional static knowledge graph to mitigate textual data biases and amplify the interpretability capabilities of the model via domain-specific dynamic knowledge graphs. The MKI distills all the knowledge and generates the final diagnostic radiology report. We performed extensive experiments on two widely used datasets, IU X-Ray and MIMIC-CXR. The experimental results demonstrate the effectiveness of our method, with all evaluation metrics outperforming previous state-of-the-art models.
Cross-domain Robust Deepfake Bias Expansion Network for Face Forgery Detection
Oct 08, 2023
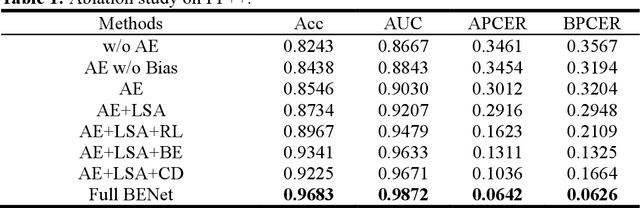
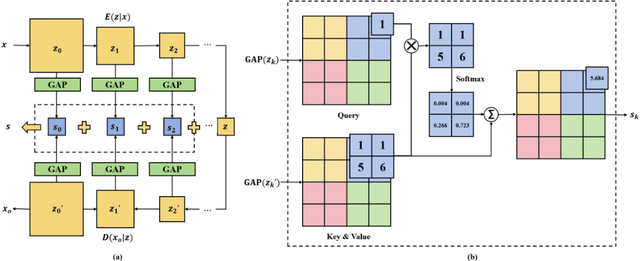
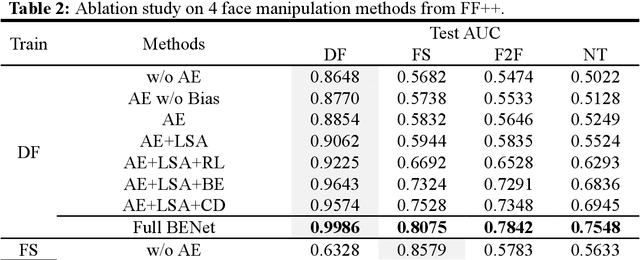
Abstract:The rapid advancement of deepfake technologies raises significant concerns about the security of face recognition systems. While existing methods leverage the clues left by deepfake techniques for face forgery detection, malicious users may intentionally manipulate forged faces to obscure the traces of deepfake clues and thereby deceive detection tools. Meanwhile, attaining cross-domain robustness for data-based methods poses a challenge due to potential gaps in the training data, which may not encompass samples from all relevant domains. Therefore, in this paper, we introduce a solution - a Cross-Domain Robust Bias Expansion Network (BENet) - designed to enhance face forgery detection. BENet employs an auto-encoder to reconstruct input faces, maintaining the invariance of real faces while selectively enhancing the difference between reconstructed fake faces and their original counterparts. This enhanced bias forms a robust foundation upon which dependable forgery detection can be built. To optimize the reconstruction results in BENet, we employ a bias expansion loss infused with contrastive concepts to attain the aforementioned objective. In addition, to further heighten the amplification of forged clues, BENet incorporates a Latent-Space Attention (LSA) module. This LSA module effectively captures variances in latent features between the auto-encoder's encoder and decoder, placing emphasis on inconsistent forgery-related information. Furthermore, BENet incorporates a cross-domain detector with a threshold to determine whether the sample belongs to a known distribution. The correction of classification results through the cross-domain detector enables BENet to defend against unknown deepfake attacks from cross-domain. Extensive experiments demonstrate the superiority of BENet compared with state-of-the-art methods in intra-database and cross-database evaluations.
Occlusion-Aware Deep Convolutional Neural Network via Homogeneous Tanh-transforms for Face Parsing
Aug 29, 2023Abstract:Face parsing infers a pixel-wise label map for each semantic facial component. Previous methods generally work well for uncovered faces, however overlook the facial occlusion and ignore some contextual area outside a single face, especially when facial occlusion has become a common situation during the COVID-19 epidemic. Inspired by the illumination theory of image, we propose a novel homogeneous tanh-transforms for image preprocessing, which made up of four tanh-transforms, that fuse the central vision and the peripheral vision together. Our proposed method addresses the dilemma of face parsing under occlusion and compresses more information of surrounding context. Based on homogeneous tanh-transforms, we propose an occlusion-aware convolutional neural network for occluded face parsing. It combines the information both in Tanh-polar space and Tanh-Cartesian space, capable of enhancing receptive fields. Furthermore, we introduce an occlusion-aware loss to focus on the boundaries of occluded regions. The network is simple and flexible, and can be trained end-to-end. To facilitate future research of occluded face parsing, we also contribute a new cleaned face parsing dataset, which is manually purified from several academic or industrial datasets, including CelebAMask-HQ, Short-video Face Parsing as well as Helen dataset and will make it public. Experiments demonstrate that our method surpasses state-of-art methods of face parsing under occlusion.
A New Computer-Aided Diagnosis System with Modified Genetic Feature Selection for BI-RADS Classification of Breast Masses in Mammograms
May 11, 2020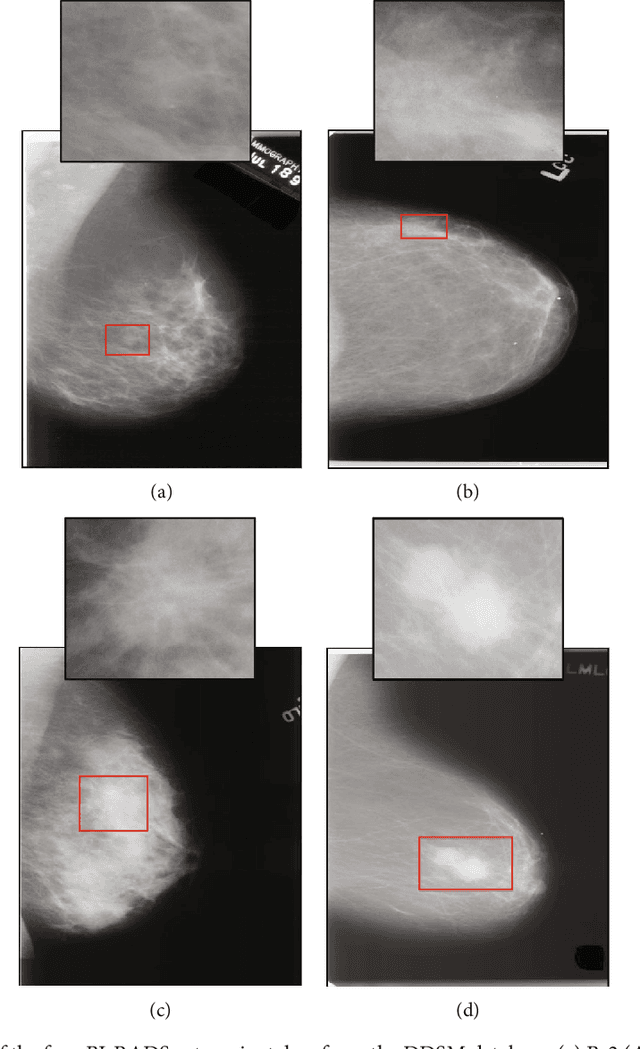
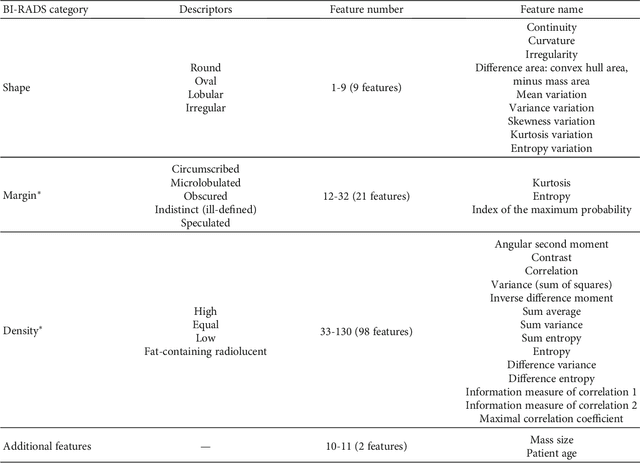
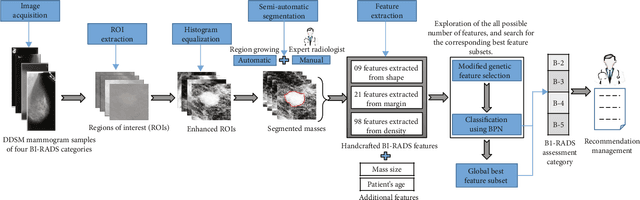
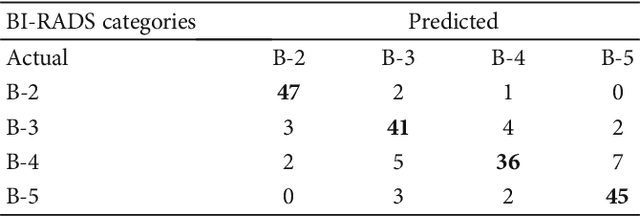
Abstract:Mammography remains the most prevalent imaging tool for early breast cancer screening. The language used to describe abnormalities in mammographic reports is based on the breast Imaging Reporting and Data System (BI-RADS). Assigning a correct BI-RADS category to each examined mammogram is a strenuous and challenging task for even experts. This paper proposes a new and effective computer-aided diagnosis (CAD) system to classify mammographic masses into four assessment categories in BI-RADS. The mass regions are first enhanced by means of histogram equalization and then semiautomatically segmented based on the region growing technique. A total of 130 handcrafted BI-RADS features are then extrcated from the shape, margin, and density of each mass, together with the mass size and the patient's age, as mentioned in BI-RADS mammography. Then, a modified feature selection method based on the genetic algorithm (GA) is proposed to select the most clinically significant BI-RADS features. Finally, a back-propagation neural network (BPN) is employed for classification, and its accuracy is used as the fitness in GA. A set of 500 mammogram images from the digital database of screening mammography (DDSM) is used for evaluation. Our system achieves classification accuracy, positive predictive value, negative predictive value, and Matthews correlation coefficient of 84.5%, 84.4%, 94.8%, and 79.3%, respectively. To our best knowledge, this is the best current result for BI-RADS classification of breast masses in mammography, which makes the proposed system promising to support radiologists for deciding proper patient management based on the automatically assigned BI-RADS categories.
A New Three-stage Curriculum Learning Approach to Deep Network Based Liver Tumor Segmentation
Oct 17, 2019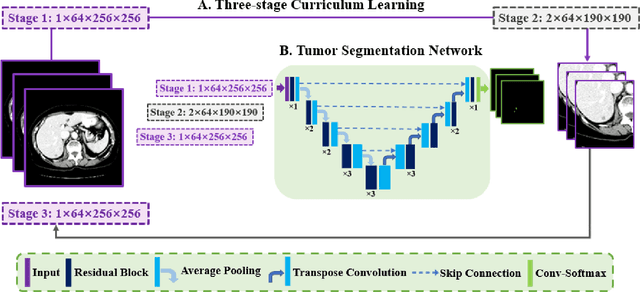
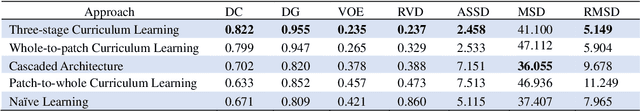
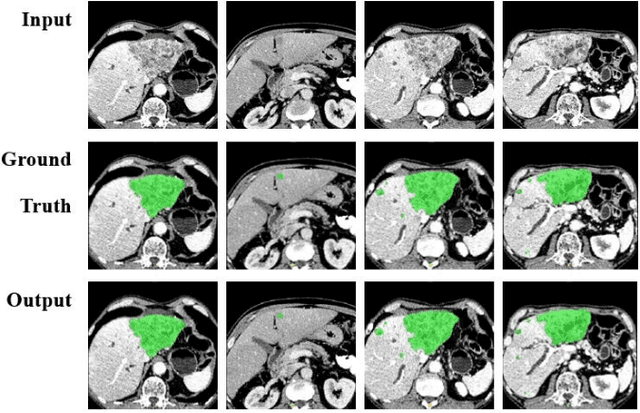
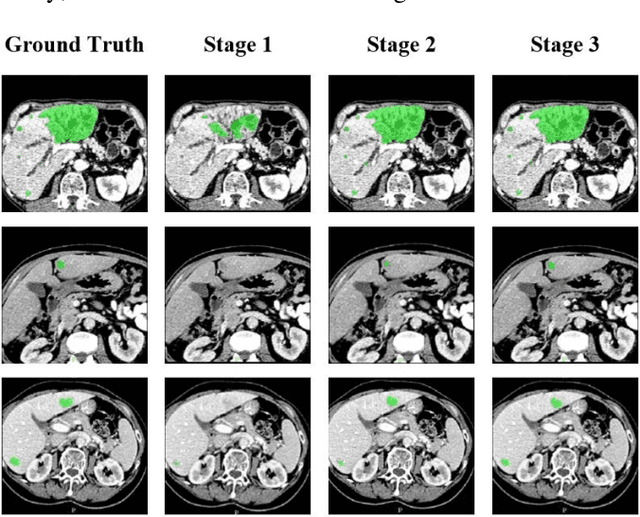
Abstract:Automatic segmentation of liver tumors in medical images is crucial for the computer-aided diagnosis and therapy. It is a challenging task, since the tumors are notoriously small against the background voxels. This paper proposes a new three-stage curriculum learning approach for training deep networks to tackle this small object segmentation problem. The learning in the first stage is performed on the whole input to obtain an initial deep network for tumor segmenta-tion. Then the second stage of learning focuses the strength-ening of tumor specific features by continuing training the network on the tumor patches. Finally, we retrain the net-work on the whole input in the third stage, in order that the tumor specific features and the global context can be inte-grated ideally under the segmentation objective. Benefitting from the proposed learning approach, we only need to em-ploy one single network to segment the tumors directly. We evaluated our approach on the 2017 MICCAI Liver Tumor Segmentation challenge dataset. In the experiments, our approach exhibits significant improvement compared with the commonly used cascaded counterpart.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge