Saad Hamid
A differentiable model of supply-chain shocks
Nov 07, 2025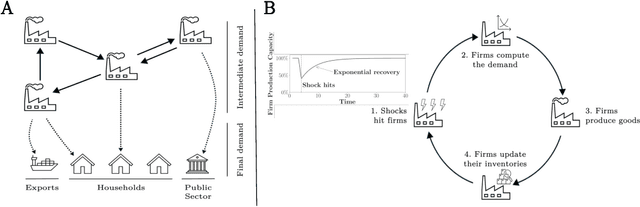
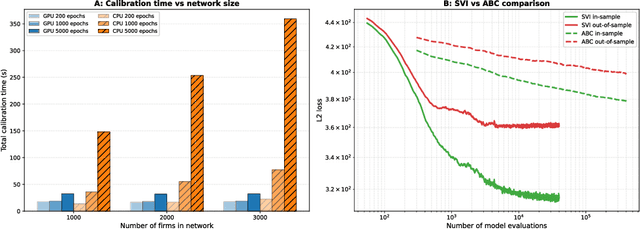
Abstract:Modelling how shocks propagate in supply chains is an increasingly important challenge in economics. Its relevance has been highlighted in recent years by events such as Covid-19 and the Russian invasion of Ukraine. Agent-based models (ABMs) are a promising approach for this problem. However, calibrating them is hard. We show empirically that it is possible to achieve speed ups of over 3 orders of magnitude when calibrating ABMs of supply networks by running them on GPUs and using automatic differentiation, compared to non-differentiable baselines. This opens the door to scaling ABMs to model the whole global supply network.
A Quadrature Approach for General-Purpose Batch Bayesian Optimization via Probabilistic Lifting
Apr 19, 2024Abstract:Parallelisation in Bayesian optimisation is a common strategy but faces several challenges: the need for flexibility in acquisition functions and kernel choices, flexibility dealing with discrete and continuous variables simultaneously, model misspecification, and lastly fast massive parallelisation. To address these challenges, we introduce a versatile and modular framework for batch Bayesian optimisation via probabilistic lifting with kernel quadrature, called SOBER, which we present as a Python library based on GPyTorch/BoTorch. Our framework offers the following unique benefits: (1) Versatility in downstream tasks under a unified approach. (2) A gradient-free sampler, which does not require the gradient of acquisition functions, offering domain-agnostic sampling (e.g., discrete and mixed variables, non-Euclidean space). (3) Flexibility in domain prior distribution. (4) Adaptive batch size (autonomous determination of the optimal batch size). (5) Robustness against a misspecified reproducing kernel Hilbert space. (6) Natural stopping criterion.
Bayesian Quadrature for Neural Ensemble Search
Mar 17, 2023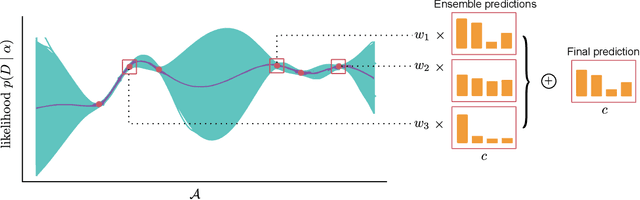
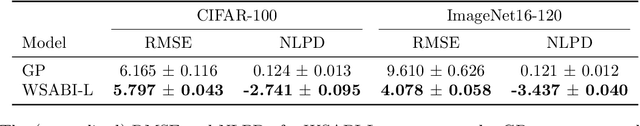
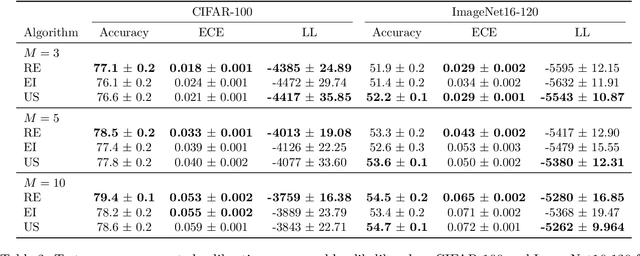
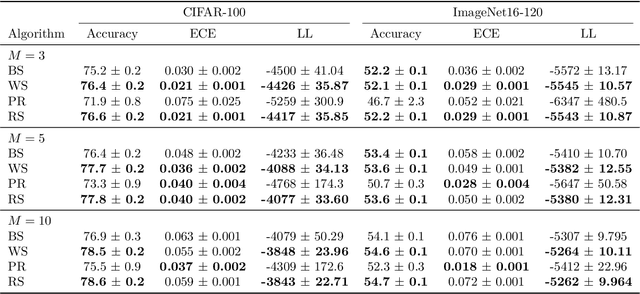
Abstract:Ensembling can improve the performance of Neural Networks, but existing approaches struggle when the architecture likelihood surface has dispersed, narrow peaks. Furthermore, existing methods construct equally weighted ensembles, and this is likely to be vulnerable to the failure modes of the weaker architectures. By viewing ensembling as approximately marginalising over architectures we construct ensembles using the tools of Bayesian Quadrature -- tools which are well suited to the exploration of likelihood surfaces with dispersed, narrow peaks. Additionally, the resulting ensembles consist of architectures weighted commensurate with their performance. We show empirically -- in terms of test likelihood, accuracy, and expected calibration error -- that our method outperforms state-of-the-art baselines, and verify via ablation studies that its components do so independently.
SOBER: Scalable Batch Bayesian Optimization and Quadrature using Recombination Constraints
Jan 30, 2023



Abstract:Batch Bayesian optimisation (BO) has shown to be a sample-efficient method of performing optimisation where expensive-to-evaluate objective functions can be queried in parallel. However, current methods do not scale to large batch sizes -- a frequent desideratum in practice (e.g. drug discovery or simulation-based inference). We present a novel algorithm, SOBER, which permits scalable and diversified batch BO with arbitrary acquisition functions, arbitrary input spaces (e.g. graph), and arbitrary kernels. The key to our approach is to reformulate batch selection for BO as a Bayesian quadrature (BQ) problem, which offers computational advantages. This reformulation is beneficial in solving BQ tasks reciprocally, which introduces the exploitative functionality of BO to BQ. We show that SOBER offers substantive performance gains in synthetic and real-world tasks, including drug discovery and simulation-based inference.
Marginalising over Stationary Kernels with Bayesian Quadrature
Jun 14, 2021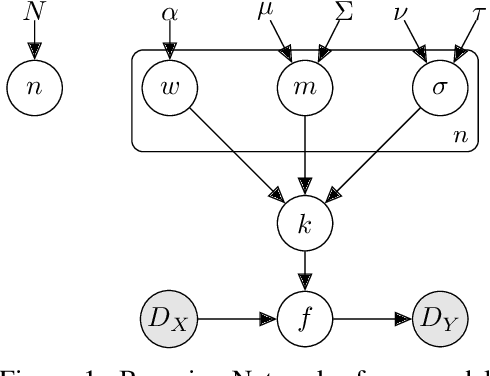

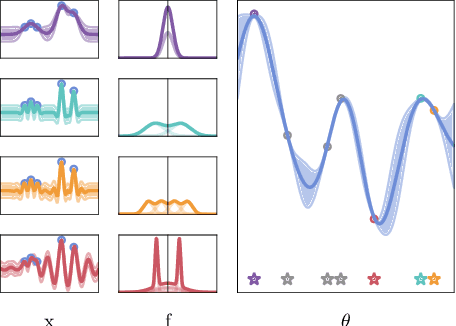

Abstract:Marginalising over families of Gaussian Process kernels produces flexible model classes with well-calibrated uncertainty estimates. Existing approaches require likelihood evaluations of many kernels, rendering them prohibitively expensive for larger datasets. We propose a Bayesian Quadrature scheme to make this marginalisation more efficient and thereby more practical. Through use of the maximum mean discrepancies between distributions, we define a kernel over kernels that captures invariances between Spectral Mixture (SM) Kernels. Kernel samples are selected by generalising an information-theoretic acquisition function for warped Bayesian Quadrature. We show that our framework achieves more accurate predictions with better calibrated uncertainty than state-of-the-art baselines, especially when given limited (wall-clock) time budgets.
Batch Selection for Parallelisation of Bayesian Quadrature
Dec 04, 2018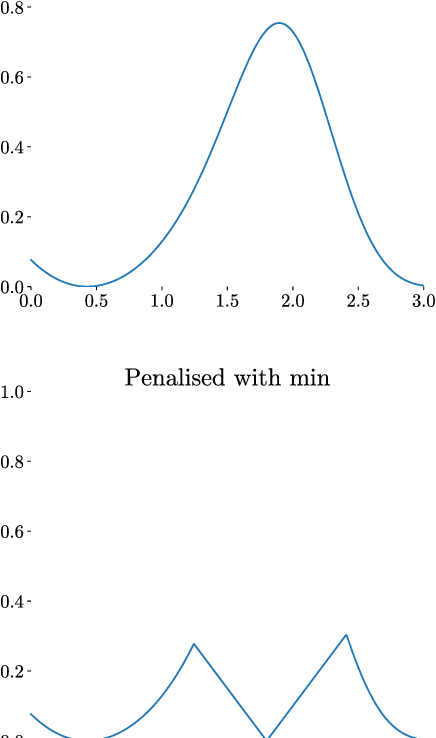
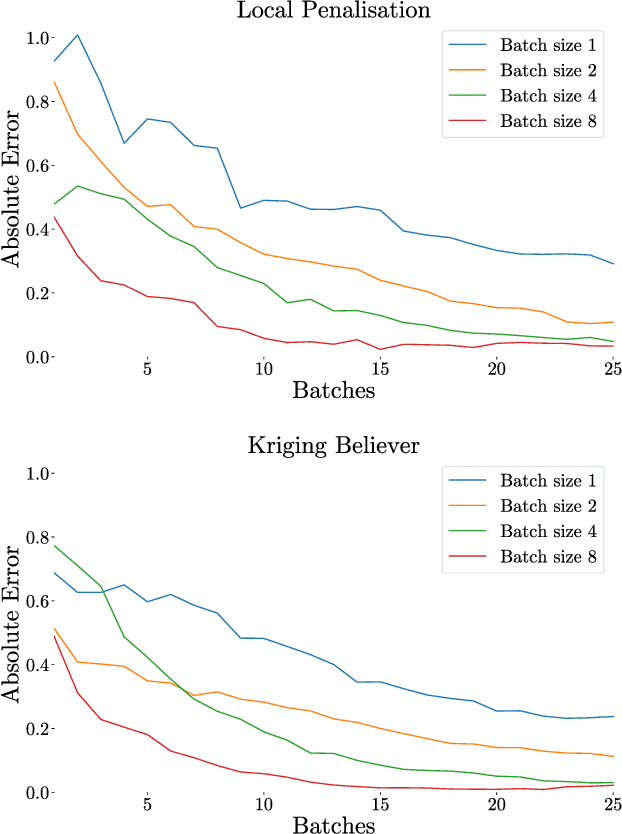
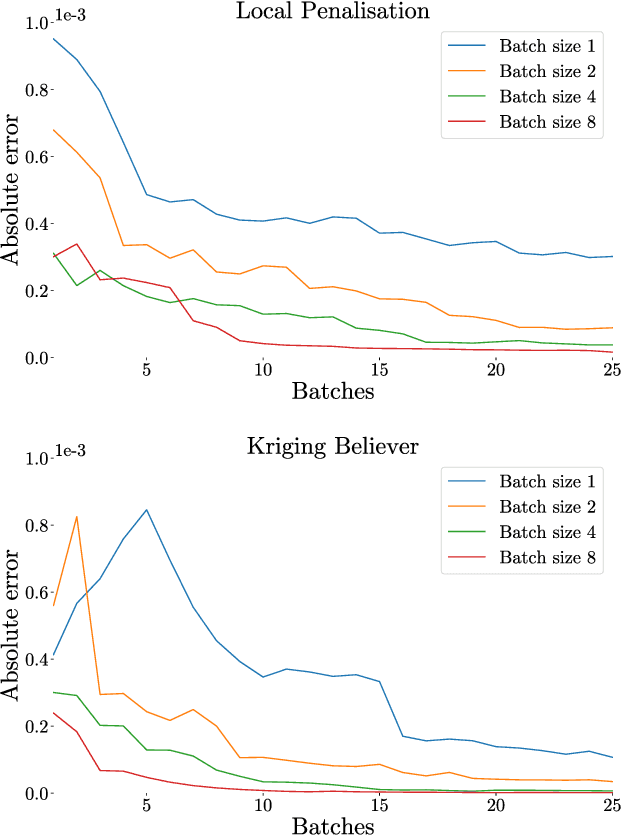
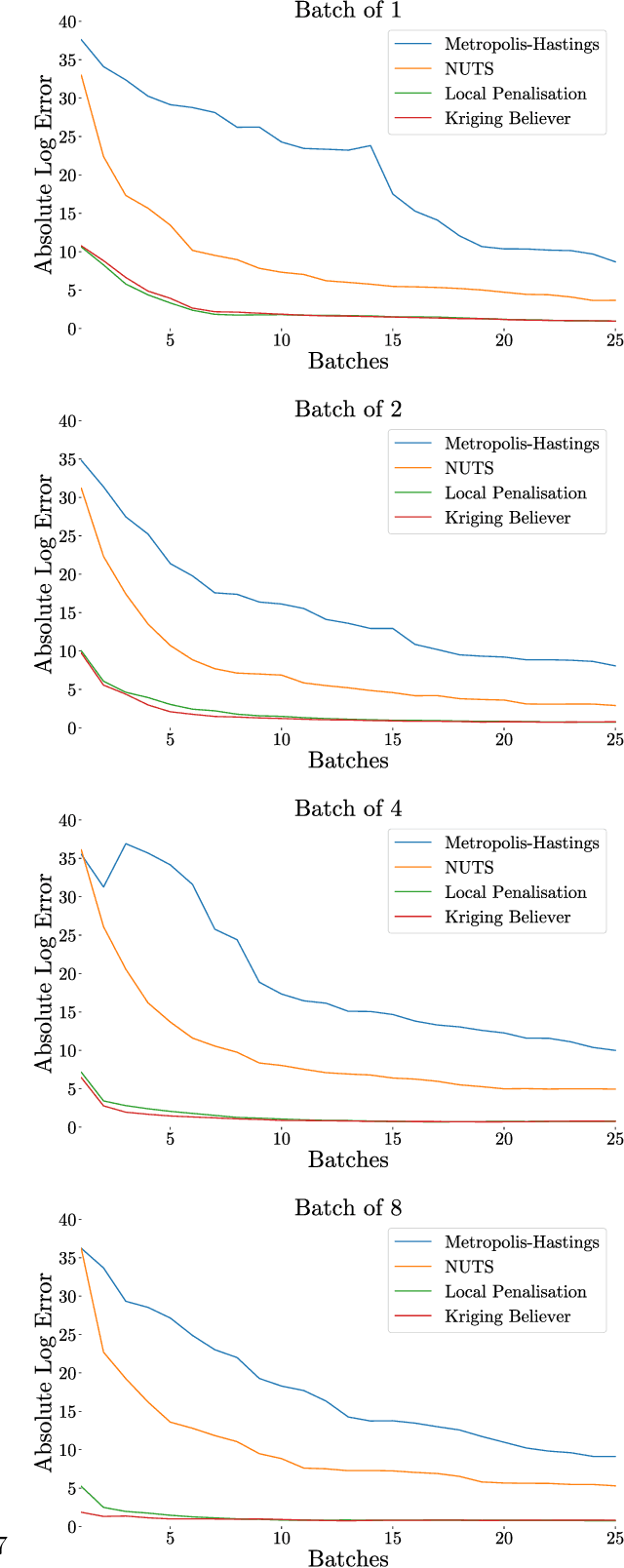
Abstract:Integration over non-negative integrands is a central problem in machine learning (e.g. for model averaging, (hyper-)parameter marginalisation, and computing posterior predictive distributions). Bayesian Quadrature is a probabilistic numerical integration technique that performs promisingly when compared to traditional Markov Chain Monte Carlo methods. However, in contrast to easily-parallelised MCMC methods, Bayesian Quadrature methods have, thus far, been essentially serial in nature, selecting a single point to sample at each step of the algorithm. We deliver methods to select batches of points at each step, based upon those recently presented in the Batch Bayesian Optimisation literature. Such parallelisation significantly reduces computation time, especially when the integrand is expensive to sample.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge