Russ B. Altman
Justice in Healthcare Artificial Intelligence in Africa
Jun 15, 2024Abstract:There is an ongoing debate on balancing the benefits and risks of artificial intelligence (AI) as AI is becoming critical to improving healthcare delivery and patient outcomes. Such improvements are essential in resource-constrained settings where millions lack access to adequate healthcare services, such as in Africa. AI in such a context can potentially improve the effectiveness, efficiency, and accessibility of healthcare services. Nevertheless, the development and use of AI-driven healthcare systems raise numerous ethical, legal, and socio-economic issues. Justice is a major concern in AI that has implications for amplifying social inequities. This paper discusses these implications and related justice concepts such as solidarity, Common Good, sustainability, AI bias, and fairness. For Africa to effectively benefit from AI, these principles should align with the local context while balancing the risks. Compared to mainstream ethical debates on justice, this perspective offers context-specific considerations for equitable healthcare AI development in Africa.
Data Ethics in the Era of Healthcare Artificial Intelligence in Africa: An Ubuntu Philosophy Perspective
Jun 14, 2024Abstract:Data are essential in developing healthcare artificial intelligence (AI) systems. However, patient data collection, access, and use raise ethical concerns, including informed consent, data bias, data protection and privacy, data ownership, and benefit sharing. Various ethical frameworks have been proposed to ensure the ethical use of healthcare data and AI, however, these frameworks often align with Western cultural values, social norms, and institutional contexts emphasizing individual autonomy and well-being. Ethical guidelines must reflect political and cultural settings to account for cultural diversity, inclusivity, and historical factors such as colonialism. Thus, this paper discusses healthcare data ethics in the AI era in Africa from the Ubuntu philosophy perspective. It focuses on the contrast between individualistic and communitarian approaches to data ethics. The proposed framework could inform stakeholders, including AI developers, healthcare providers, the public, and policy-makers about healthcare data ethical usage in AI in Africa.
Detecting Contradictory COVID-19 Drug Efficacy Claims from Biomedical Literature
Dec 19, 2022



Abstract:The COVID-19 pandemic created a deluge of questionable and contradictory scientific claims about drug efficacy -- an "infodemic" with lasting consequences for science and society. In this work, we argue that NLP models can help domain experts distill and understand the literature in this complex, high-stakes area. Our task is to automatically identify contradictory claims about COVID-19 drug efficacy. We frame this as a natural language inference problem and offer a new NLI dataset created by domain experts. The NLI framing allows us to create curricula combining existing datasets and our own. The resulting models are useful investigative tools. We provide a case study of how these models help a domain expert summarize and assess evidence concerning remdisivir and hydroxychloroquine.
POPDx: An Automated Framework for Patient Phenotyping across 392,246 Individuals in the UK Biobank Study
Aug 23, 2022



Abstract:Objective For the UK Biobank standardized phenotype codes are associated with patients who have been hospitalized but are missing for many patients who have been treated exclusively in an outpatient setting. We describe a method for phenotype recognition that imputes phenotype codes for all UK Biobank participants. Materials and Methods POPDx (Population-based Objective Phenotyping by Deep Extrapolation) is a bilinear machine learning framework for simultaneously estimating the probabilities of 1,538 phenotype codes. We extracted phenotypic and health-related information of 392,246 individuals from the UK Biobank for POPDx development and evaluation. A total of 12,803 ICD-10 diagnosis codes of the patients were converted to 1,538 Phecodes as gold standard labels. The POPDx framework was evaluated and compared to other available methods on automated multi-phenotype recognition. Results POPDx can predict phenotypes that are rare or even unobserved in training. We demonstrate substantial improvement of automated multi-phenotype recognition across 22 disease categories, and its application in identifying key epidemiological features associated with each phenotype. Conclusions POPDx helps provide well-defined cohorts for downstream studies. It is a general purpose method that can be applied to other biobanks with diverse but incomplete data.
ATOM3D: Tasks On Molecules in Three Dimensions
Dec 07, 2020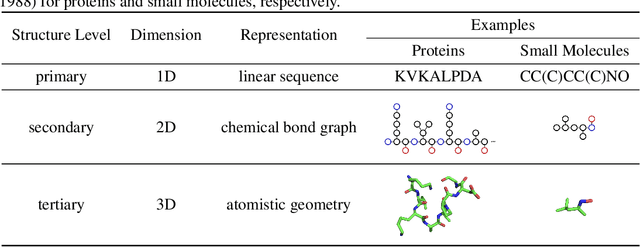
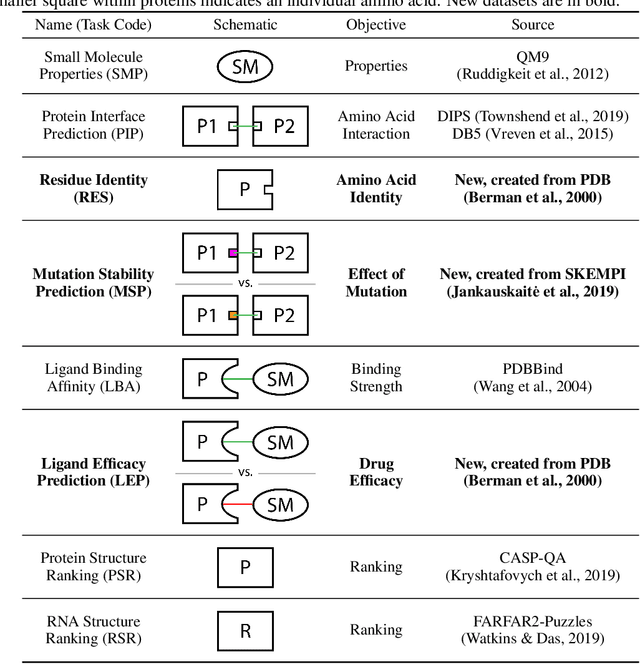
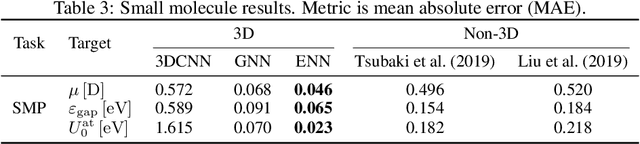
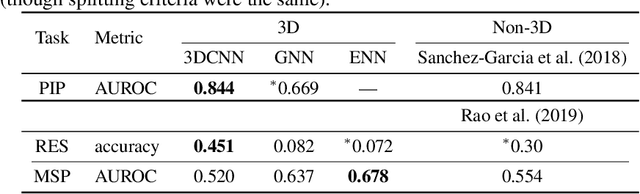
Abstract:Computational methods that operate directly on three-dimensional molecular structure hold large potential to solve important questions in biology and chemistry. In particular deep neural networks have recently gained significant attention. In this work we present ATOM3D, a collection of both novel and existing datasets spanning several key classes of biomolecules, to systematically assess such learning methods. We develop three-dimensional molecular learning networks for each of these tasks, finding that they consistently improve performance relative to one- and two-dimensional methods. The specific choice of architecture proves to be critical for performance, with three-dimensional convolutional networks excelling at tasks involving complex geometries, while graph networks perform well on systems requiring detailed positional information. Furthermore, equivariant networks show significant promise. Our results indicate many molecular problems stand to gain from three-dimensional molecular learning. All code and datasets can be accessed via https://www.atom3d.ai .
A Probabilistic Algorithm for Calculating Structure: Borrowing from Simulated Annealing
Mar 06, 2013



Abstract:We have developed a general Bayesian algorithm for determining the coordinates of points in a three-dimensional space. The algorithm takes as input a set of probabilistic constraints on the coordinates of the points, and an a priori distribution for each point location. The output is a maximum-likelihood estimate of the location of each point. We use the extended, iterated Kalman filter, and add a search heuristic for optimizing its solution under nonlinear conditions. This heuristic is based on the same principle as the simulated annealing heuristic for other optimization problems. Our method uses any probabilistic constraints that can be expressed as a function of the point coordinates (for example, distance, angles, dihedral angles, and planarity). It assumes that all constraints have Gaussian noise. In this paper, we describe the algorithm and show its performance on a set of synthetic data to illustrate its convergence properties, and its applicability to domains such ng molecular structure determination.
Probabilistic Constraint Satisfaction with Non-Gaussian Noise
Feb 27, 2013


Abstract:We have previously reported a Bayesian algorithm for determining the coordinates of points in three-dimensional space from uncertain constraints. This method is useful in the determination of biological molecular structure. It is limited, however, by the requirement that the uncertainty in the constraints be normally distributed. In this paper, we present an extension of the original algorithm that allows constraint uncertainty to be represented as a mixture of Gaussians, and thereby allows arbitrary constraint distributions. We illustrate the performance of this algorithm on a problem drawn from the domain of molecular structure determination, in which a multicomponent constraint representation produces a much more accurate solution than the old single component mechanism. The new mechanism uses mixture distributions to decompose the problem into a set of independent problems with unimodal constraint uncertainty. The results of the unimodal subproblems are periodically recombined using Bayes' law, to avoid combinatorial explosion. The new algorithm is particularly suited for parallel implementation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge