Robert Mendel
Regensburg Medical Image Computing
Motion-Corrected Moving Average: Including Post-Hoc Temporal Information for Improved Video Segmentation
Mar 05, 2024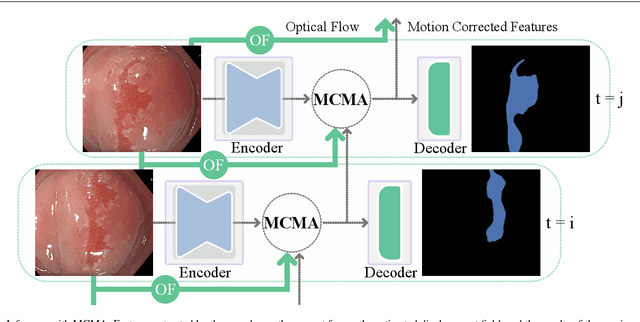
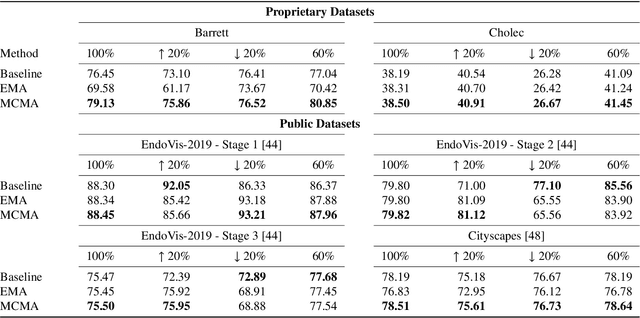
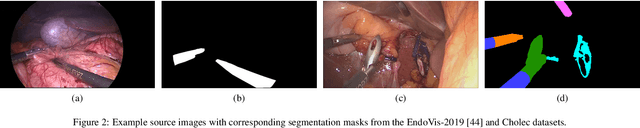
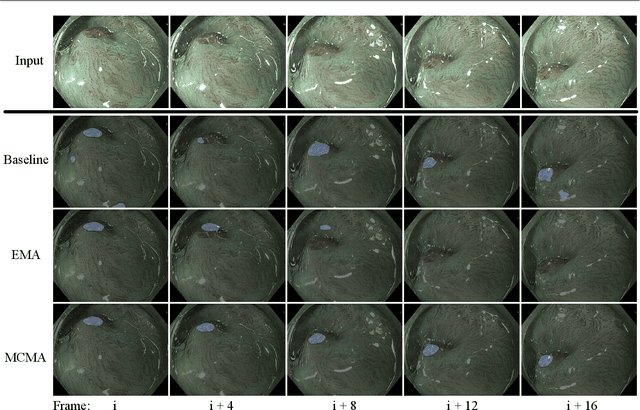
Abstract:Real-time computational speed and a high degree of precision are requirements for computer-assisted interventions. Applying a segmentation network to a medical video processing task can introduce significant inter-frame prediction noise. Existing approaches can reduce inconsistencies by including temporal information but often impose requirements on the architecture or dataset. This paper proposes a method to include temporal information in any segmentation model and, thus, a technique to improve video segmentation performance without alterations during training or additional labeling. With Motion-Corrected Moving Average, we refine the exponential moving average between the current and previous predictions. Using optical flow to estimate the movement between consecutive frames, we can shift the prior term in the moving-average calculation to align with the geometry of the current frame. The optical flow calculation does not require the output of the model and can therefore be performed in parallel, leading to no significant runtime penalty for our approach. We evaluate our approach on two publicly available segmentation datasets and two proprietary endoscopic datasets and show improvements over a baseline approach.
Learning Visual Representations with Optimum-Path Forest and its Applications to Barrett's Esophagus and Adenocarcinoma Diagnosis
Jan 19, 2021
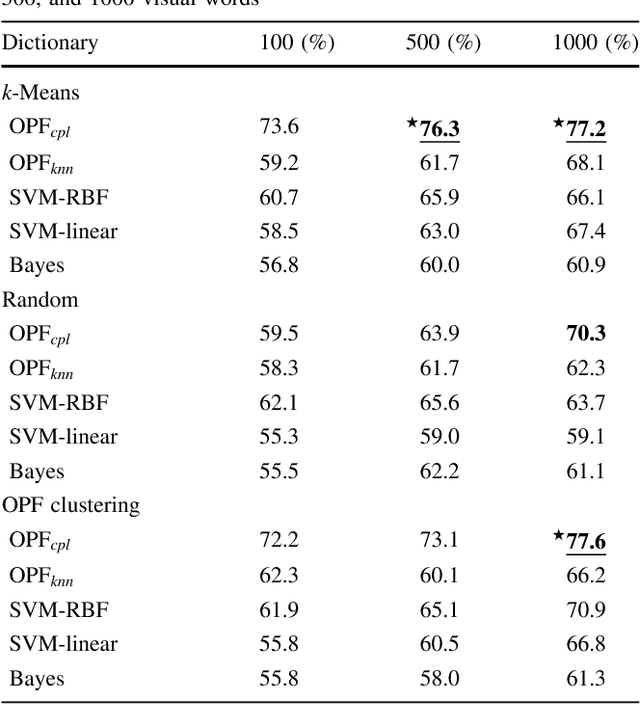
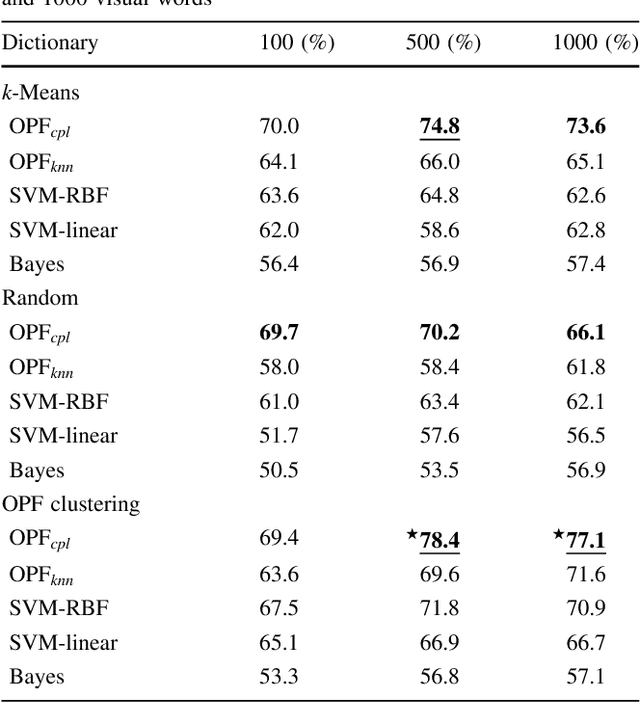
Abstract:In this work, we introduce the unsupervised Optimum-Path Forest (OPF) classifier for learning visual dictionaries in the context of Barrett's esophagus (BE) and automatic adenocarcinoma diagnosis. The proposed approach was validated in two datasets (MICCAI 2015 and Augsburg) using three different feature extractors (SIFT, SURF, and the not yet applied to the BE context A-KAZE), as well as five supervised classifiers, including two variants of the OPF, Support Vector Machines with Radial Basis Function and Linear kernels, and a Bayesian classifier. Concerning MICCAI 2015 dataset, the best results were obtained using unsupervised OPF for dictionary generation using supervised OPF for classification purposes and using SURF feature extractor with accuracy nearly to 78% for distinguishing BE patients from adenocarcinoma ones. Regarding the Augsburg dataset, the most accurate results were also obtained using both OPF classifiers but with A-KAZE as the feature extractor with accuracy close to 73%. The combination of feature extraction and bag-of-visual-words techniques showed results that outperformed others obtained recently in the literature, as well as we highlight new advances in the related research area. Reinforcing the significance of this work, to the best of our knowledge, this is the first one that aimed at addressing computer-aided BE identification using bag-of-visual-words and OPF classifiers, being this application of unsupervised technique in the BE feature calculation the major contribution of this work. It is also proposed a new BE and adenocarcinoma description using the A-KAZE features, not yet applied in the literature.
2018 Robotic Scene Segmentation Challenge
Feb 03, 2020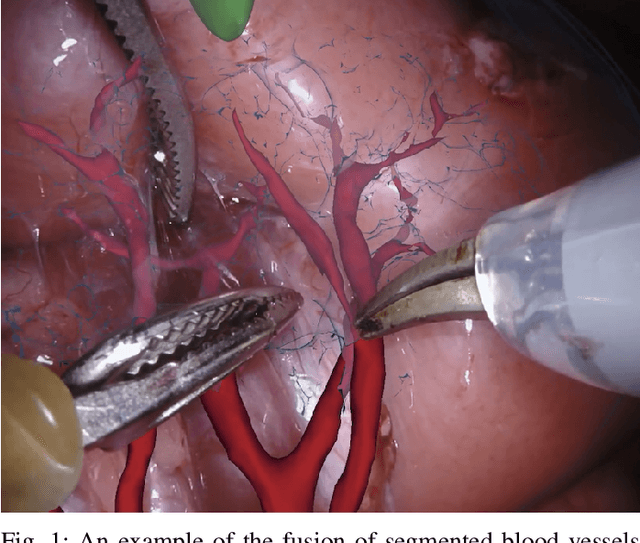
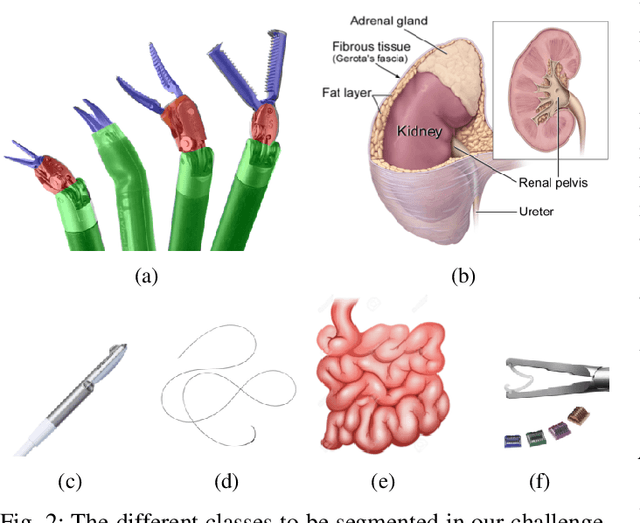
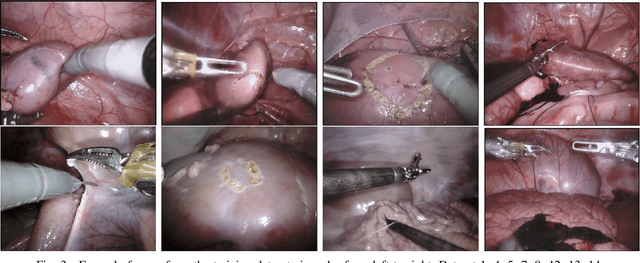
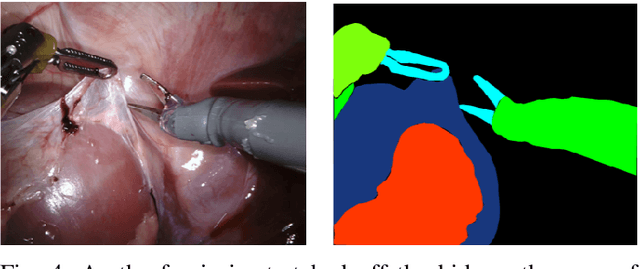
Abstract:In 2015 we began a sub-challenge at the EndoVis workshop at MICCAI in Munich using endoscope images of ex-vivo tissue with automatically generated annotations from robot forward kinematics and instrument CAD models. However, the limited background variation and simple motion rendered the dataset uninformative in learning about which techniques would be suitable for segmentation in real surgery. In 2017, at the same workshop in Quebec we introduced the robotic instrument segmentation dataset with 10 teams participating in the challenge to perform binary, articulating parts and type segmentation of da Vinci instruments. This challenge included realistic instrument motion and more complex porcine tissue as background and was widely addressed with modifications on U-Nets and other popular CNN architectures. In 2018 we added to the complexity by introducing a set of anatomical objects and medical devices to the segmented classes. To avoid over-complicating the challenge, we continued with porcine data which is dramatically simpler than human tissue due to the lack of fatty tissue occluding many organs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge