Phattarapong Sawangjai
PseudoCell: Hard Negative Mining as Pseudo Labeling for Deep Learning-Based Centroblast Cell Detection
Jul 06, 2023Abstract:Patch classification models based on deep learning have been utilized in whole-slide images (WSI) of H&E-stained tissue samples to assist pathologists in grading follicular lymphoma patients. However, these approaches still require pathologists to manually identify centroblast cells and provide refined labels for optimal performance. To address this, we propose PseudoCell, an object detection framework to automate centroblast detection in WSI (source code is available at https://github.com/IoBT-VISTEC/PseudoCell.git). This framework incorporates centroblast labels from pathologists and combines them with pseudo-negative labels obtained from undersampled false-positive predictions using the cell's morphological features. By employing PseudoCell, pathologists' workload can be reduced as it accurately narrows down the areas requiring their attention during examining tissue. Depending on the confidence threshold, PseudoCell can eliminate 58.18-99.35% of non-centroblasts tissue areas on WSI. This study presents a practical centroblast prescreening method that does not require pathologists' refined labels for improvement. Detailed guidance on the practical implementation of PseudoCell is provided in the discussion section.
EEG-BBNet: a Hybrid Framework for Brain Biometric using Graph Connectivity
Aug 17, 2022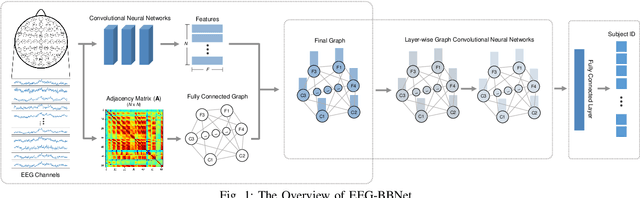
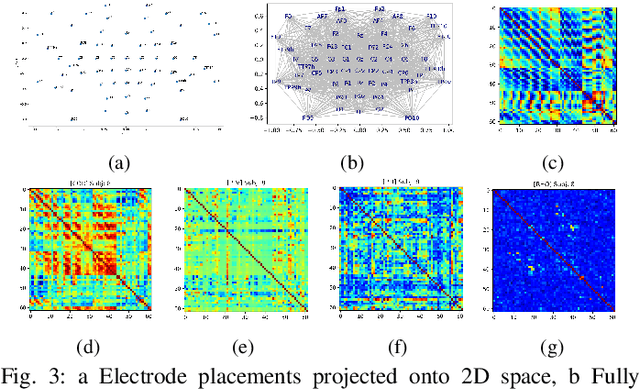
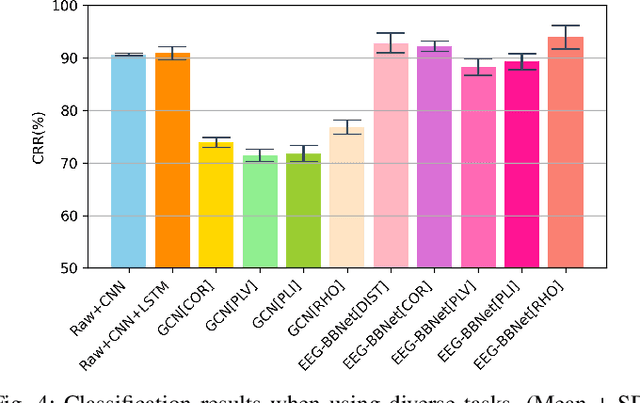
Abstract:Brain biometrics based on electroencephalography (EEG) have been used increasingly for personal identification. Traditional machine learning techniques as well as modern day deep learning methods have been applied with promising results. In this paper we present EEG-BBNet, a hybrid network which integrates convolutional neural networks (CNN) with graph convolutional neural networks (GCNN). The benefit of the CNN in automatic feature extraction and the capability of GCNN in learning connectivity between EEG electrodes through graph representation are jointly exploited. We examine various connectivity measures, namely the Euclidean distance, Pearson's correlation coefficient, phase-locked value, phase-lag index, and Rho index. The performance of the proposed method is assessed on a benchmark dataset consisting of various brain-computer interface (BCI) tasks and compared to other state-of-the-art approaches. We found that our models outperform all baselines in the event-related potential (ERP) task with an average correct recognition rates up to 99.26% using intra-session data. EEG-BBNet with Pearson's correlation and RHO index provide the best classification results. In addition, our model demonstrates greater adaptability using inter-session and inter-task data. We also investigate the practicality of our proposed model with smaller number of electrodes. Electrode placements over the frontal lobe region appears to be most appropriate with minimal lost in performance.
OCTAve: 2D en face Optical Coherence Tomography Angiography Vessel Segmentation in Weakly-Supervised Learning with Locality Augmentation
Jul 25, 2022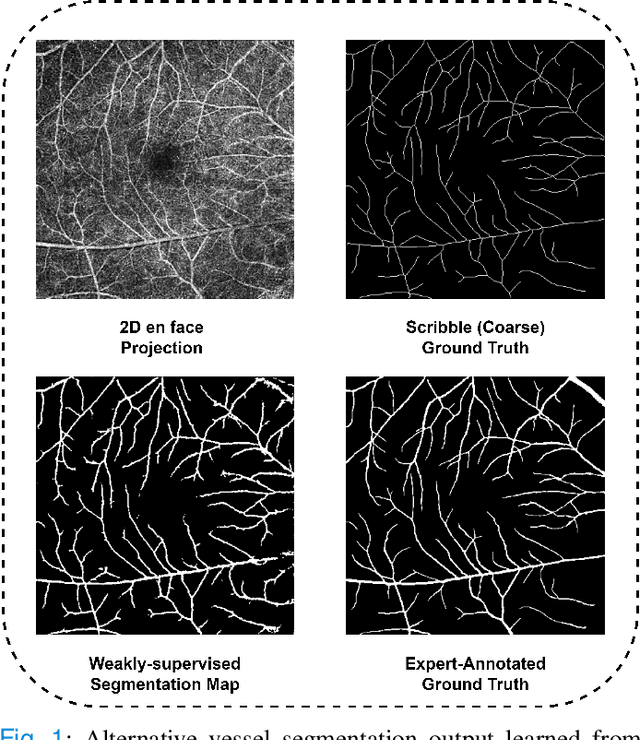
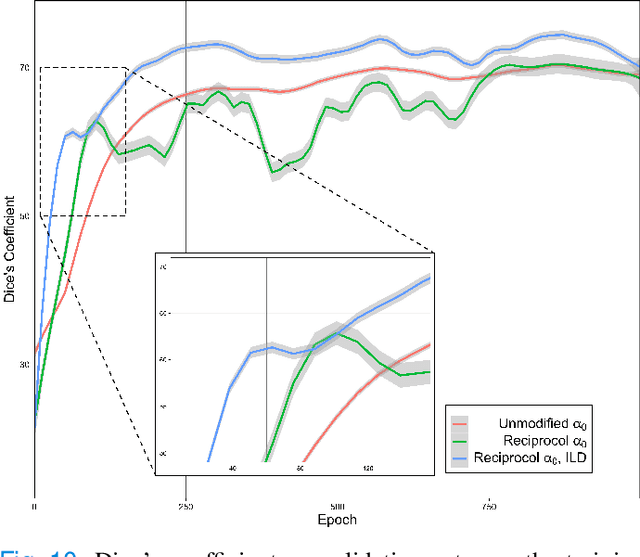

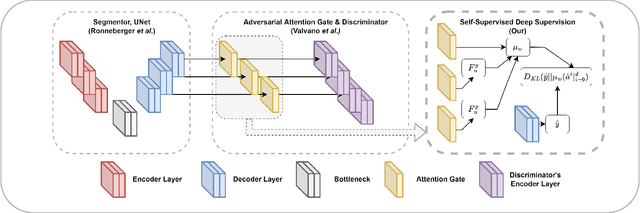
Abstract:While there have been increased researches using deep learning techniques for the extraction of vascular structure from the 2D en face OCTA, for such approach, it is known that the data annotation process on the curvilinear structure like the retinal vasculature is very costly and time consuming, albeit few tried to address the annotation problem. In this work, we propose the application of the scribble-base weakly-supervised learning method to automate the pixel-level annotation. The proposed method, called OCTAve, combines the weakly-supervised learning using scribble-annotated ground truth augmented with an adversarial and a novel self-supervised deep supervision. Our novel mechanism is designed to utilize the discriminative outputs from the discrimination layer of a UNet-like architecture where the Kullback-Liebler Divergence between the aggregate discriminative outputs and the segmentation map predicate is minimized during the training. This combined method leads to the better localization of the vascular structure as shown in our experiments. We validate our proposed method on the large public datasets i.e., ROSE, OCTA-500. The segmentation performance is compared against both state-of-the-art fully-supervised and scribble-based weakly-supervised approaches. The implementation of our work used in the experiments is located at [LINK].
A comparative study for interpreting deep learning prediction of the Parkinson's disease diagnosis from SPECT imaging
Aug 23, 2019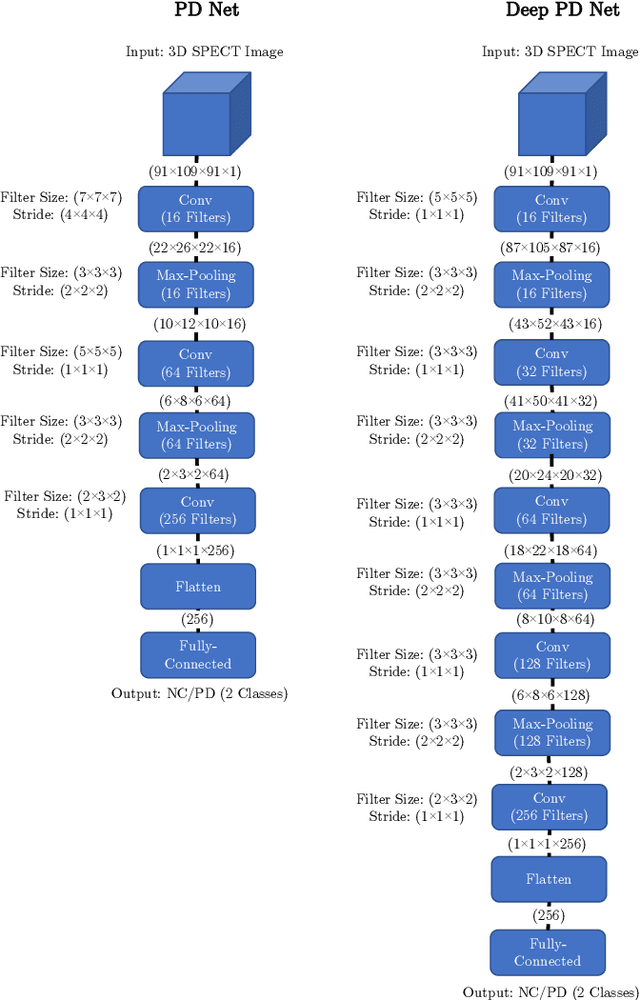
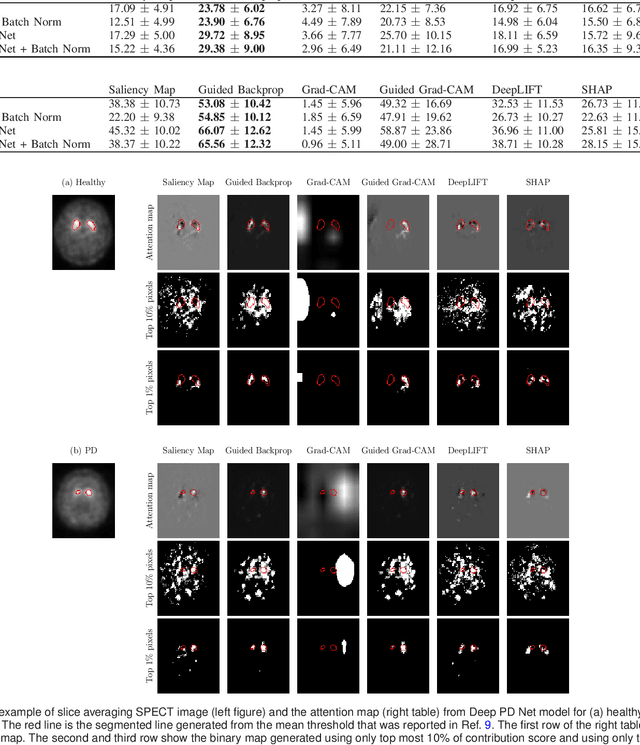
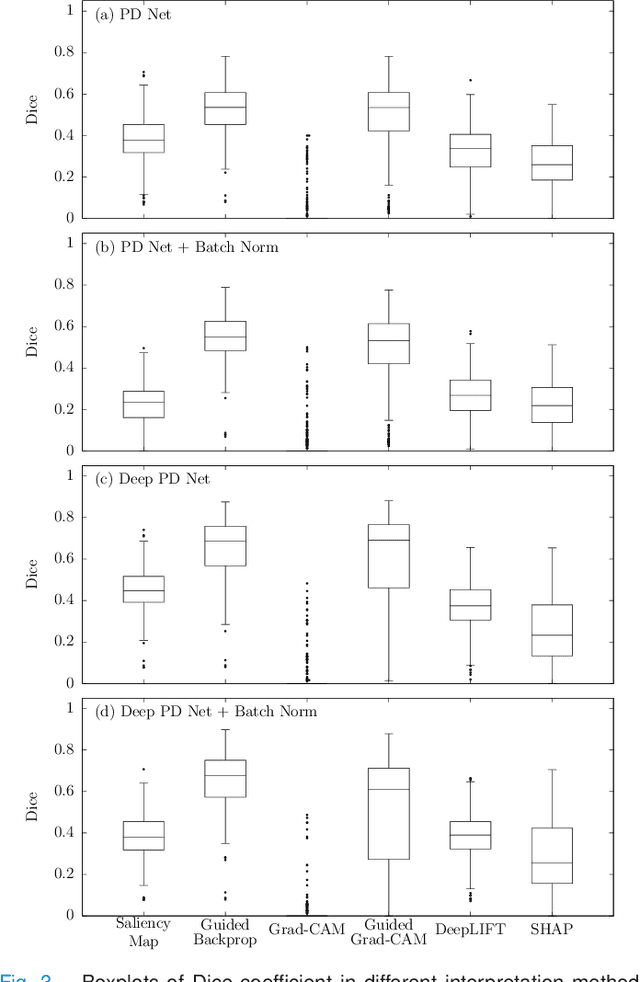

Abstract:The application of deep learning to single-photon emission computed tomography (SPECT) imaging in Parkinson's disease shows effectively high diagnosis accuracy. However, difficulties in model interpretation were occurred due to the complexity of the deep learning model. Although several interpretation methods were created to show the attention map that contains important features of the input data, it is still uncertain whether these methods can be applied in PD diagnosis. Four different models of the deep learning approach based on 3-dimensional convolution neural network (3D-CNN) of well-established architectures have been trained with an accuracy up to 95-96% in classification performance. These four models have been used as the comparative study for well-known interpretation methods. Generally, radiologists interpret SPECT images by confirming the shape of the I123-Ioflupane uptake in the striatal nuclei. To evaluate the interpretation performance, the segmented striatal nuclei of SPECT images are chosen as the ground truth. Results suggest that guided backpropagation and SHAP which were developed recently, provided the best interpretation performance. Guided backpropagation has the best performance to generate the attention map that focuses on the location of striatal nuclei. On the other hand, SHAP surpasses other methods in suggesting the change of the striatal nucleus uptake shape from healthy to PD subjects. Results from both methods confirm that 3D-CNN focuses on the striatal nuclei in the same way as the radiologist, and both methods should be suggested to increase the credibility of the model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge