Sanparith Marukatat
Automatic Detection of Rice Disease in Images of Various Leaf Sizes
Jun 15, 2022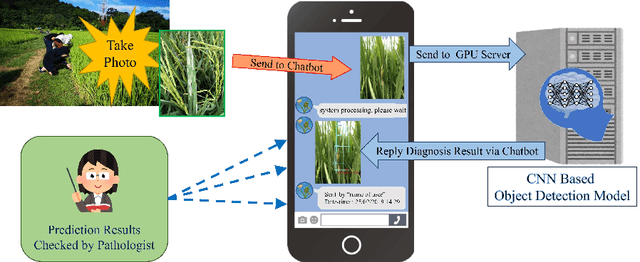
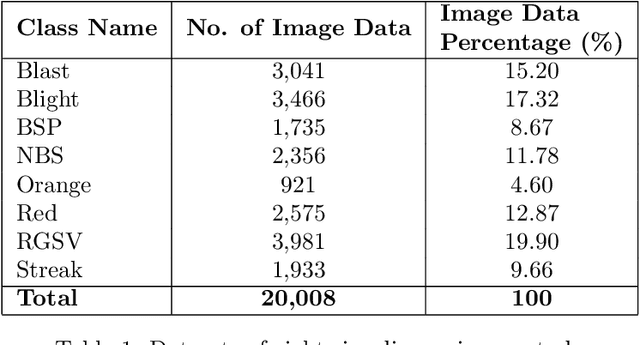
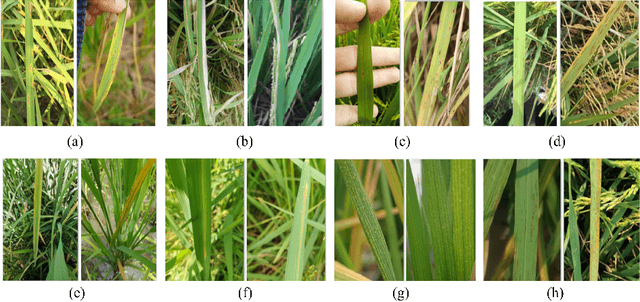
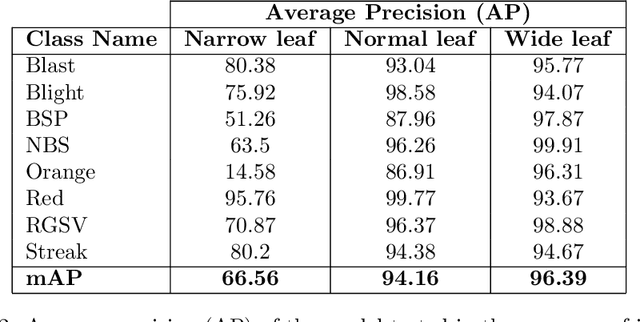
Abstract:Fast, accurate and affordable rice disease detection method is required to assist rice farmers tackling equipment and expertise shortages problems. In this paper, we focused on the solution using computer vision technique to detect rice diseases from rice field photograph images. Dealing with images took in real-usage situation by general farmers is quite challenging due to various environmental factors, and rice leaf object size variation is one major factor caused performance gradation. To solve this problem, we presented a technique combining a CNN object detection with image tiling technique, based on automatically estimated width size of rice leaves in the images as a size reference for dividing the original input image. A model to estimate leaf width was created by small size CNN such as 18 layer ResNet architecture model. A new divided tiled sub-image set with uniformly sized object was generated and used as input for training a rice disease prediction model. Our technique was evaluated on 4,960 images of eight different types of rice leaf diseases, including blast, blight, brown spot, narrow brown spot, orange, red stripe, rice grassy stunt virus, and streak disease. The mean absolute percentage error (MAPE) for leaf width prediction task evaluated on all eight classes was 11.18% in the experiment, indicating that the leaf width prediction model performed well. The mean average precision (mAP) of the prediction performance on YOLOv4 architecture was enhanced from 87.56% to 91.14% when trained and tested with the tiled dataset. According to our study, the proposed image tiling technique improved rice disease detection efficiency.
A comparative study for interpreting deep learning prediction of the Parkinson's disease diagnosis from SPECT imaging
Aug 23, 2019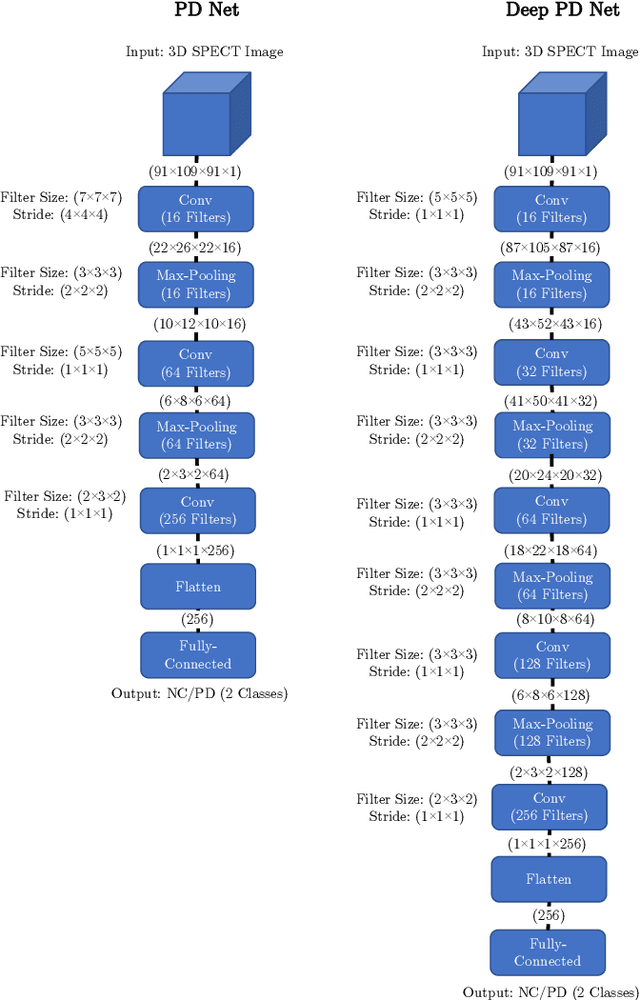
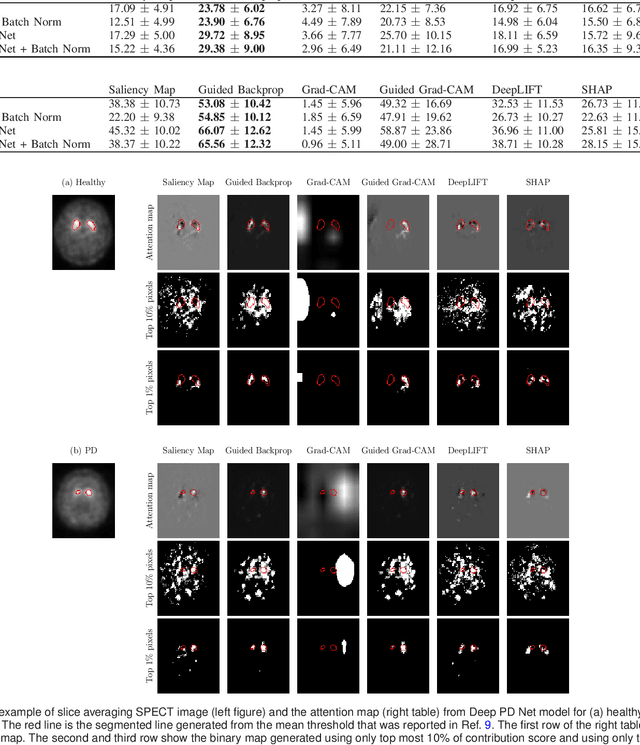
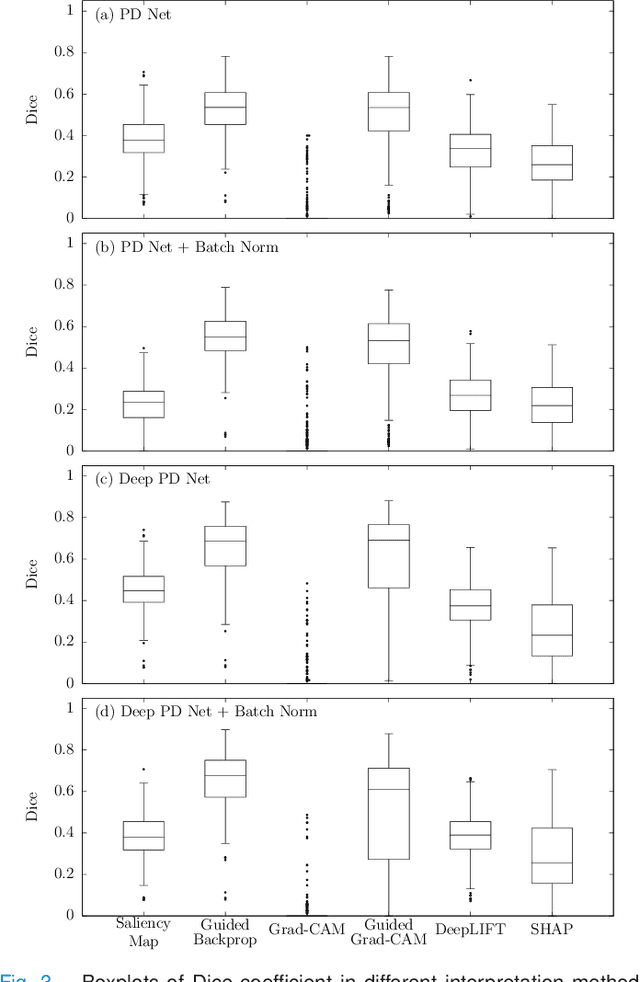

Abstract:The application of deep learning to single-photon emission computed tomography (SPECT) imaging in Parkinson's disease shows effectively high diagnosis accuracy. However, difficulties in model interpretation were occurred due to the complexity of the deep learning model. Although several interpretation methods were created to show the attention map that contains important features of the input data, it is still uncertain whether these methods can be applied in PD diagnosis. Four different models of the deep learning approach based on 3-dimensional convolution neural network (3D-CNN) of well-established architectures have been trained with an accuracy up to 95-96% in classification performance. These four models have been used as the comparative study for well-known interpretation methods. Generally, radiologists interpret SPECT images by confirming the shape of the I123-Ioflupane uptake in the striatal nuclei. To evaluate the interpretation performance, the segmented striatal nuclei of SPECT images are chosen as the ground truth. Results suggest that guided backpropagation and SHAP which were developed recently, provided the best interpretation performance. Guided backpropagation has the best performance to generate the attention map that focuses on the location of striatal nuclei. On the other hand, SHAP surpasses other methods in suggesting the change of the striatal nucleus uptake shape from healthy to PD subjects. Results from both methods confirm that 3D-CNN focuses on the striatal nuclei in the same way as the radiologist, and both methods should be suggested to increase the credibility of the model.
Automatic Lung Cancer Prediction from Chest X-ray Images Using Deep Learning Approach
Aug 31, 2018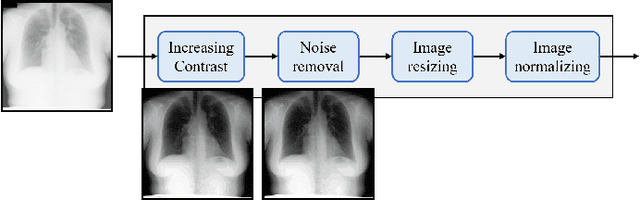
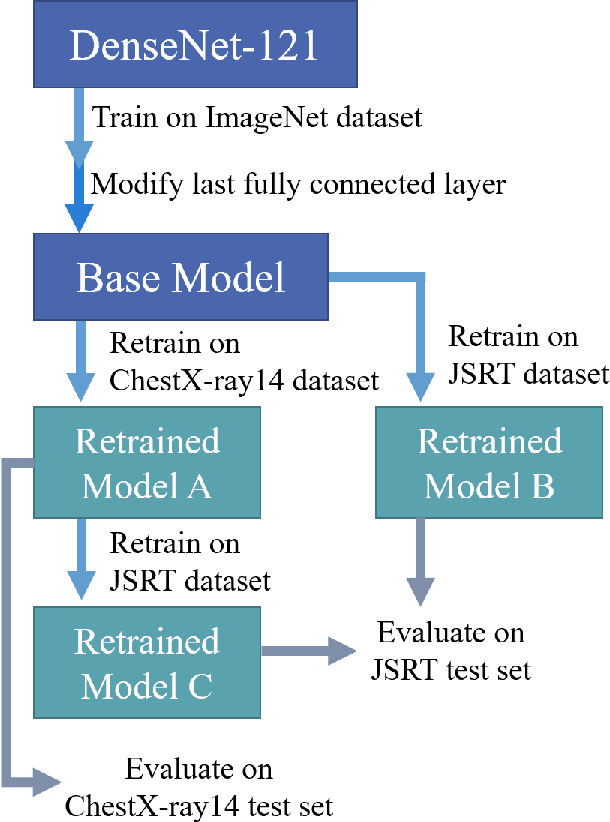
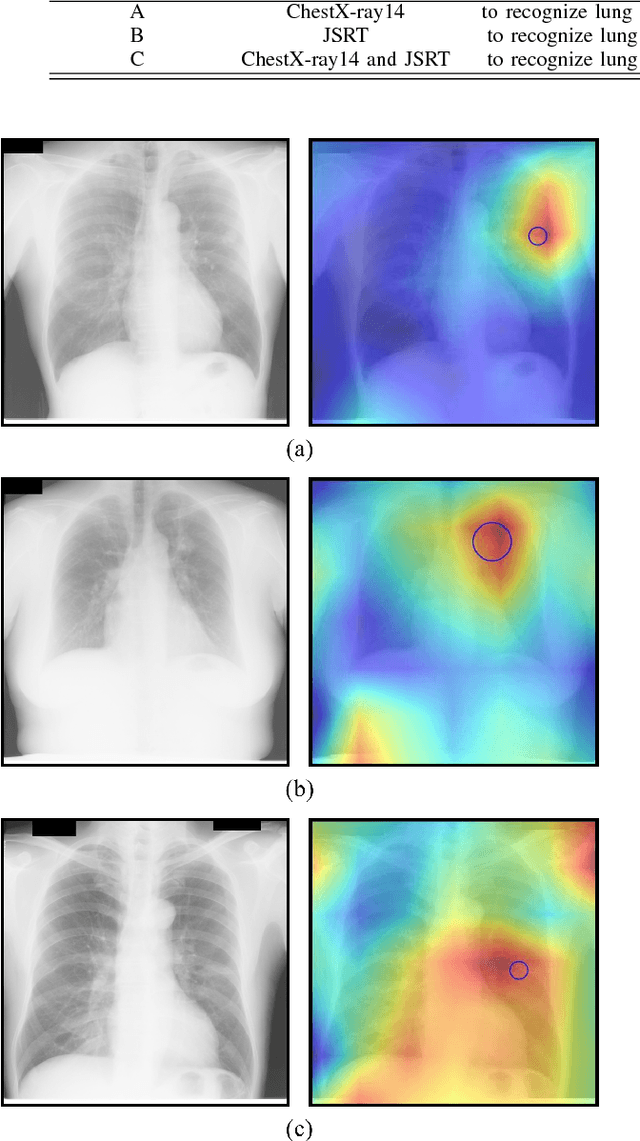
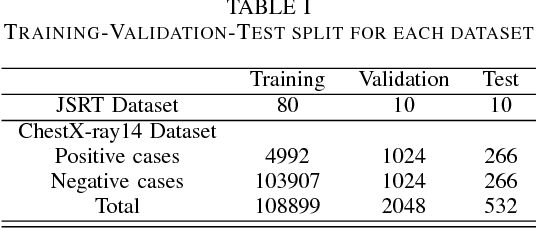
Abstract:Since, cancer is curable when diagnosed at an early stage, lung cancer screening plays an important role in preventive care. Although both low dose computed tomography (LDCT) and computed tomography (CT) scans provide more medical information than normal chest x-rays, there is very limited access to these technologies in rural areas. Recently, there is a trend in using computer-aided diagnosis (CADx) to assist in screening and diagnosing of cancer from biomedical images. In this study, the 121-layer convolutional neural network also known as DenseNet-121 by G. Huang et. al., along with the transfer learning scheme was explored as a means to classify lung cancer using chest X-ray images. The model was trained on a lung nodules dataset before training on the lung cancer dataset to alleviate the problem of a small dataset. The proposed model yields 74.43$\pm$6.01\% of mean accuracy, 74.96$\pm$9.85\% of mean specificity, and 74.68$\pm$15.33\% of mean sensitivity. The proposed model also provides a heatmap for identifying the location of the lung nodule. These findings are promising for further development of chest x-ray-based lung cancer diagnosis using the deep learning approach. Moreover, these findings solve the problem of small dataset.
Counterfactual Mean Embedding: A Kernel Method for Nonparametric Causal Inference
May 22, 2018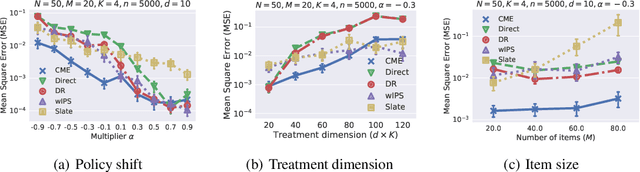
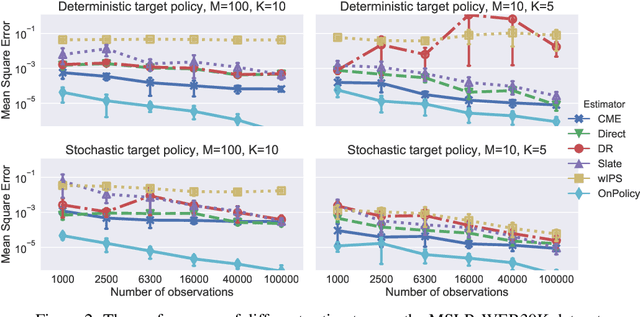
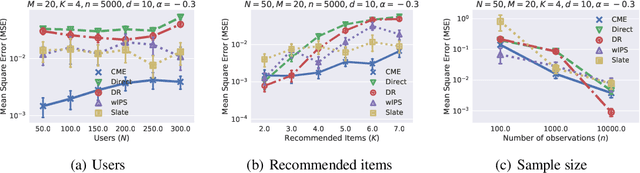
Abstract:This paper introduces a novel Hilbert space representation of a counterfactual distribution---called counterfactual mean embedding (CME)---with applications in nonparametric causal inference. Counterfactual prediction has become an ubiquitous tool in machine learning applications, such as online advertisement, recommendation systems, and medical diagnosis, whose performance relies on certain interventions. To infer the outcomes of such interventions, we propose to embed the associated counterfactual distribution into a reproducing kernel Hilbert space (RKHS) endowed with a positive definite kernel. Under appropriate assumptions, the CME allows us to perform causal inference over the entire landscape of the counterfactual distribution. The CME can be estimated consistently from observational data without requiring any parametric assumption about the underlying distributions. We also derive a rate of convergence which depends on the smoothness of the conditional mean and the Radon-Nikodym derivative of the underlying marginal distributions. Our framework can deal with not only real-valued outcome, but potentially also more complex and structured outcomes such as images, sequences, and graphs. Lastly, our experimental results on off-policy evaluation tasks demonstrate the advantages of the proposed estimator.
Automatic Training Data Synthesis for Handwriting Recognition Using the Structural Crossing-Over Technique
Oct 09, 2014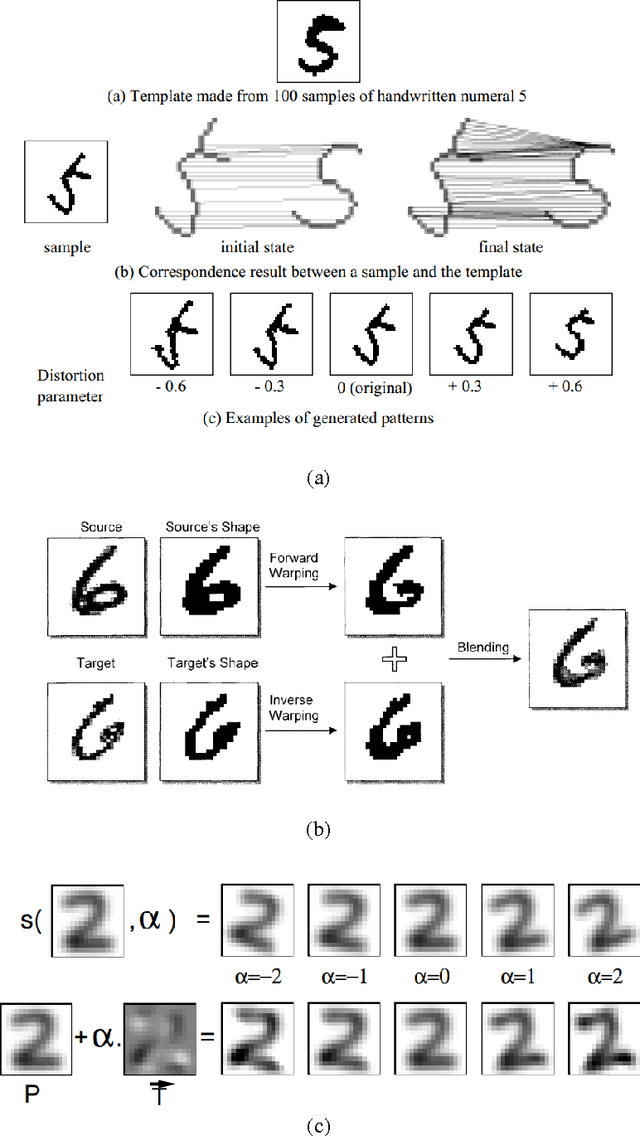

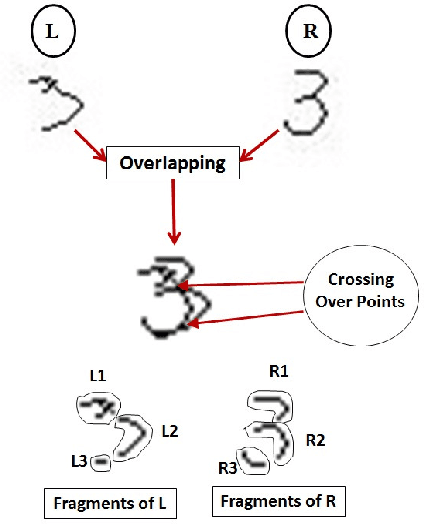
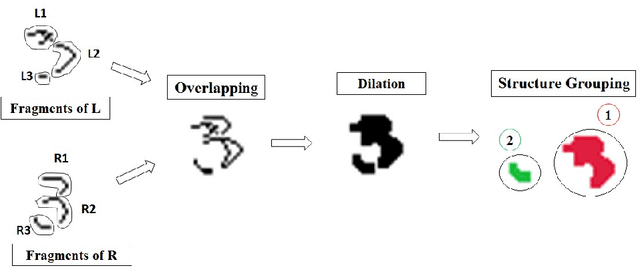
Abstract:The paper presents a novel technique called "Structural Crossing-Over" to synthesize qualified data for training machine learning-based handwriting recognition. The proposed technique can provide a greater variety of patterns of training data than the existing approaches such as elastic distortion and tangent-based affine transformation. A couple of training characters are chosen, then they are analyzed by their similar and different structures, and finally are crossed over to generate the new characters. The experiments are set to compare the performances of tangent-based affine transformation and the proposed approach in terms of the variety of generated characters and percent of recognition errors. The standard MNIST corpus including 60,000 training characters and 10,000 test characters is employed in the experiments. The proposed technique uses 1,000 characters to synthesize 60,000 characters, and then uses these data to train and test the benchmark handwriting recognition system that exploits Histogram of Gradient (HOG) as features and Support Vector Machine (SVM) as recognizer. The experimental result yields 8.06% of errors. It significantly outperforms the tangent-based affine transformation and the original MNIST training data, which are 11.74% and 16.55%, respectively.
* 8 pages, 6 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge