Noman Mohammed
Privacy-Preserving Fair Synthetic Tabular Data
Mar 04, 2025Abstract:Sharing of tabular data containing valuable but private information is limited due to legal and ethical issues. Synthetic data could be an alternative solution to this sharing problem, as it is artificially generated by machine learning algorithms and tries to capture the underlying data distribution. However, machine learning models are not free from memorization and may introduce biases, as they rely on training data. Producing synthetic data that preserves privacy and fairness while maintaining utility close to the real data is a challenging task. This research simultaneously addresses both the privacy and fairness aspects of synthetic data, an area not explored by other studies. In this work, we present PF-WGAN, a privacy-preserving, fair synthetic tabular data generator based on the WGAN-GP model. We have modified the original WGAN-GP by adding privacy and fairness constraints forcing it to produce privacy-preserving fair data. This approach will enable the publication of datasets that protect individual's privacy and remain unbiased toward any particular group. We compared the results with three state-of-the-art synthetic data generator models in terms of utility, privacy, and fairness across four different datasets. We found that the proposed model exhibits a more balanced trade-off among utility, privacy, and fairness.
Robust Privacy Amidst Innovation with Large Language Models Through a Critical Assessment of the Risks
Jul 23, 2024


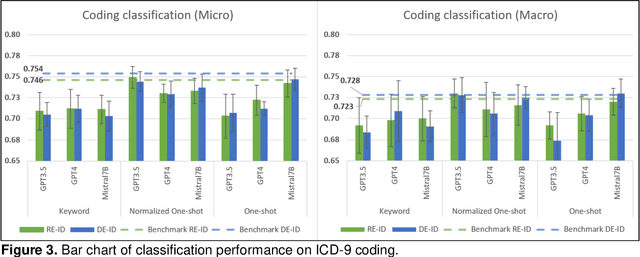
Abstract:This study examines integrating EHRs and NLP with large language models (LLMs) to improve healthcare data management and patient care. It focuses on using advanced models to create secure, HIPAA-compliant synthetic patient notes for biomedical research. The study used de-identified and re-identified MIMIC III datasets with GPT-3.5, GPT-4, and Mistral 7B to generate synthetic notes. Text generation employed templates and keyword extraction for contextually relevant notes, with one-shot generation for comparison. Privacy assessment checked PHI occurrence, while text utility was tested using an ICD-9 coding task. Text quality was evaluated with ROUGE and cosine similarity metrics to measure semantic similarity with source notes. Analysis of PHI occurrence and text utility via the ICD-9 coding task showed that the keyword-based method had low risk and good performance. One-shot generation showed the highest PHI exposure and PHI co-occurrence, especially in geographic location and date categories. The Normalized One-shot method achieved the highest classification accuracy. Privacy analysis revealed a critical balance between data utility and privacy protection, influencing future data use and sharing. Re-identified data consistently outperformed de-identified data. This study demonstrates the effectiveness of keyword-based methods in generating privacy-protecting synthetic clinical notes that retain data usability, potentially transforming clinical data-sharing practices. The superior performance of re-identified over de-identified data suggests a shift towards methods that enhance utility and privacy by using dummy PHIs to perplex privacy attacks.
Synthetic Data: Revisiting the Privacy-Utility Trade-off
Jul 09, 2024Abstract:Synthetic data has been considered a better privacy-preserving alternative to traditionally sanitized data across various applications. However, a recent article challenges this notion, stating that synthetic data does not provide a better trade-off between privacy and utility than traditional anonymization techniques, and that it leads to unpredictable utility loss and highly unpredictable privacy gain. The article also claims to have identified a breach in the differential privacy guarantees provided by PATEGAN and PrivBayes. When a study claims to refute or invalidate prior findings, it is crucial to verify and validate the study. In our work, we analyzed the implementation of the privacy game described in the article and found that it operated in a highly specialized and constrained environment, which limits the applicability of its findings to general cases. Our exploration also revealed that the game did not satisfy a crucial precondition concerning data distributions, which contributed to the perceived violation of the differential privacy guarantees offered by PATEGAN and PrivBayes. We also conducted a privacy-utility trade-off analysis in a more general and unconstrained environment. Our experimentation demonstrated that synthetic data achieves a more favorable privacy-utility trade-off compared to the provided implementation of k-anonymization, thereby reaffirming earlier conclusions.
De-identification is not always enough
Jan 31, 2024Abstract:For sharing privacy-sensitive data, de-identification is commonly regarded as adequate for safeguarding privacy. Synthetic data is also being considered as a privacy-preserving alternative. Recent successes with numerical and tabular data generative models and the breakthroughs in large generative language models raise the question of whether synthetically generated clinical notes could be a viable alternative to real notes for research purposes. In this work, we demonstrated that (i) de-identification of real clinical notes does not protect records against a membership inference attack, (ii) proposed a novel approach to generate synthetic clinical notes using the current state-of-the-art large language models, (iii) evaluated the performance of the synthetically generated notes in a clinical domain task, and (iv) proposed a way to mount a membership inference attack where the target model is trained with synthetic data. We observed that when synthetically generated notes closely match the performance of real data, they also exhibit similar privacy concerns to the real data. Whether other approaches to synthetically generated clinical notes could offer better trade-offs and become a better alternative to sensitive real notes warrants further investigation.
Federated Learning Algorithms for Generalized Mixed-effects Model (GLMM) on Horizontally Partitioned Data from Distributed Sources
Sep 28, 2021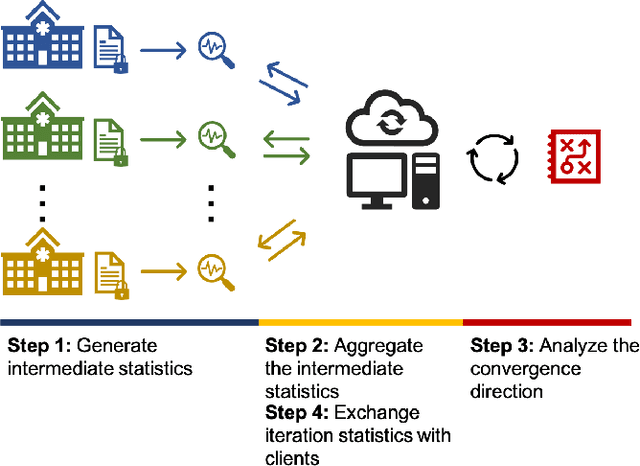

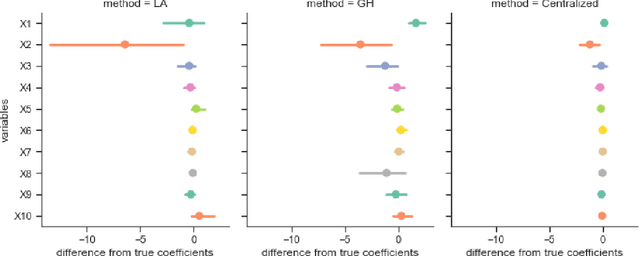
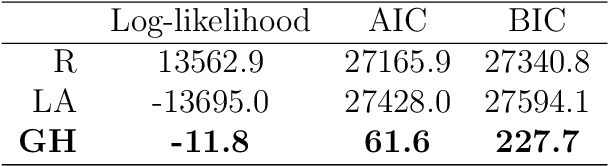
Abstract:Objectives: This paper develops two algorithms to achieve federated generalized linear mixed effect models (GLMM), and compares the developed model's outcomes with each other, as well as that from the standard R package (`lme4'). Methods: The log-likelihood function of GLMM is approximated by two numerical methods (Laplace approximation and Gaussian Hermite approximation), which supports federated decomposition of GLMM to bring computation to data. Results: Our developed method can handle GLMM to accommodate hierarchical data with multiple non-independent levels of observations in a federated setting. The experiment results demonstrate comparable (Laplace) and superior (Gaussian-Hermite) performances with simulated and real-world data. Conclusion: We developed and compared federated GLMMs with different approximations, which can support researchers in analyzing biomedical data to accommodate mixed effects and address non-independence due to hierarchical structures (i.e., institutes, region, country, etc.).
De-identification of Unstructured Clinical Texts from Sequence to Sequence Perspective
Sep 10, 2021

Abstract:In this work, we propose a novel problem formulation for de-identification of unstructured clinical text. We formulate the de-identification problem as a sequence to sequence learning problem instead of a token classification problem. Our approach is inspired by the recent state-of -the-art performance of sequence to sequence learning models for named entity recognition. Early experimentation of our proposed approach achieved 98.91% recall rate on i2b2 dataset. This performance is comparable to current state-of-the-art models for unstructured clinical text de-identification.
Feature Selection for Classification under Anonymity Constraint
Feb 06, 2017



Abstract:Over the last decade, proliferation of various online platforms and their increasing adoption by billions of users have heightened the privacy risk of a user enormously. In fact, security researchers have shown that sparse microdata containing information about online activities of a user although anonymous, can still be used to disclose the identity of the user by cross-referencing the data with other data sources. To preserve the privacy of a user, in existing works several methods (k-anonymity, l-diversity, differential privacy) are proposed that ensure a dataset which is meant to share or publish bears small identity disclosure risk. However, the majority of these methods modify the data in isolation, without considering their utility in subsequent knowledge discovery tasks, which makes these datasets less informative. In this work, we consider labeled data that are generally used for classification, and propose two methods for feature selection considering two goals: first, on the reduced feature set the data has small disclosure risk, and second, the utility of the data is preserved for performing a classification task. Experimental results on various real-world datasets show that the method is effective and useful in practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge