Nico Goernitz
SynsetRank: Degree-adjusted Random Walk for Relation Identification
Sep 15, 2016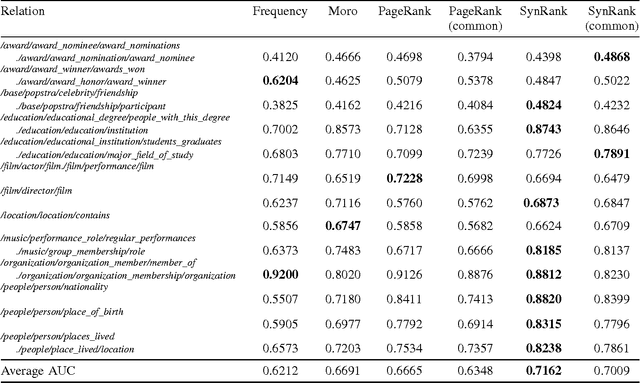

Abstract:In relation extraction, a key process is to obtain good detectors that find relevant sentences describing the target relation. To minimize the necessity of labeled data for refining detectors, previous work successfully made use of BabelNet, a semantic graph structure expressing relationships between synsets, as side information or prior knowledge. The goal of this paper is to enhance the use of graph structure in the framework of random walk with a few adjustable parameters. Actually, a straightforward application of random walk degrades the performance even after parameter optimization. With the insight from this unsuccessful trial, we propose SynsetRank, which adjusts the initial probability so that high degree nodes influence the neighbors as strong as low degree nodes. In our experiment on 13 relations in the FB15K-237 dataset, SynsetRank significantly outperforms baselines and the plain random walk approach.
Toward Supervised Anomaly Detection
Jan 23, 2014



Abstract:Anomaly detection is being regarded as an unsupervised learning task as anomalies stem from adversarial or unlikely events with unknown distributions. However, the predictive performance of purely unsupervised anomaly detection often fails to match the required detection rates in many tasks and there exists a need for labeled data to guide the model generation. Our first contribution shows that classical semi-supervised approaches, originating from a supervised classifier, are inappropriate and hardly detect new and unknown anomalies. We argue that semi-supervised anomaly detection needs to ground on the unsupervised learning paradigm and devise a novel algorithm that meets this requirement. Although being intrinsically non-convex, we further show that the optimization problem has a convex equivalent under relatively mild assumptions. Additionally, we propose an active learning strategy to automatically filter candidates for labeling. In an empirical study on network intrusion detection data, we observe that the proposed learning methodology requires much less labeled data than the state-of-the-art, while achieving higher detection accuracies.
mTim: Rapid and accurate transcript reconstruction from RNA-Seq data
Sep 20, 2013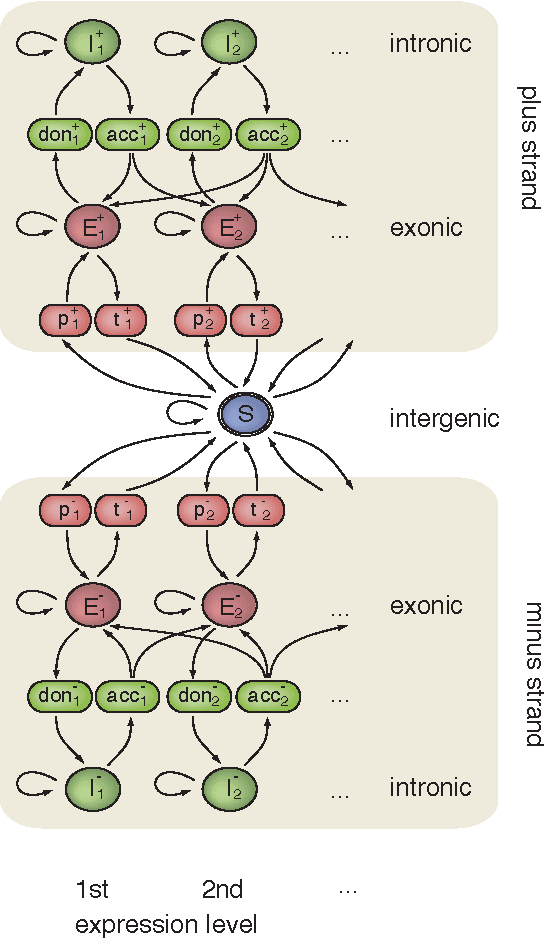
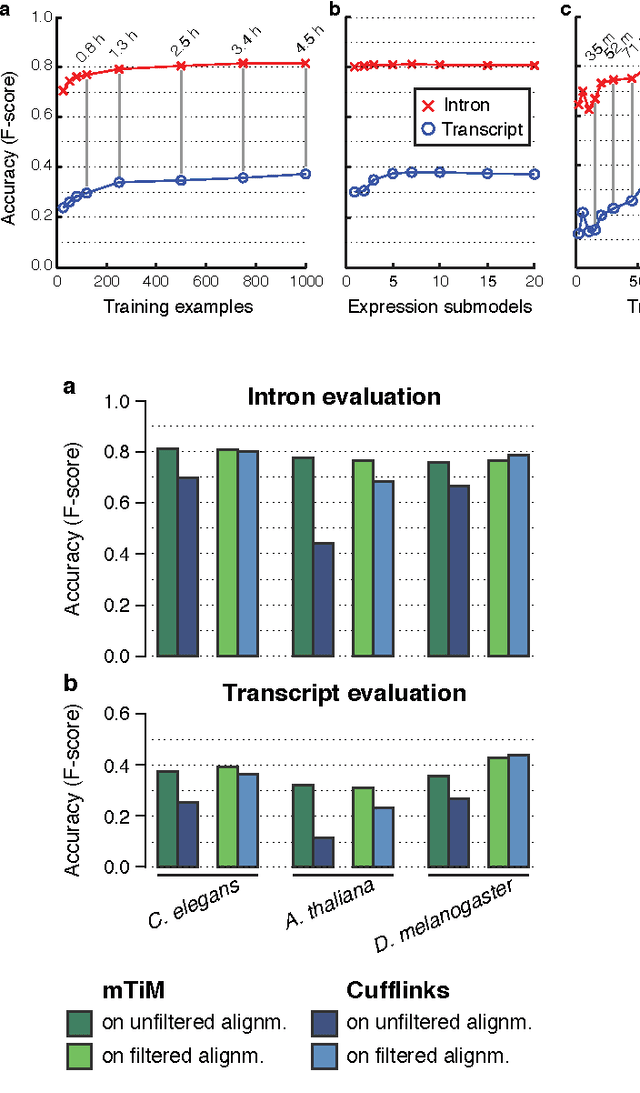
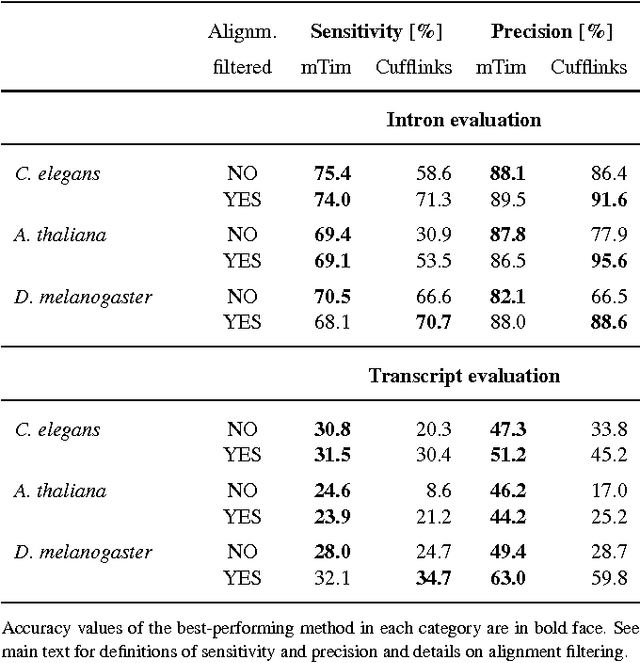
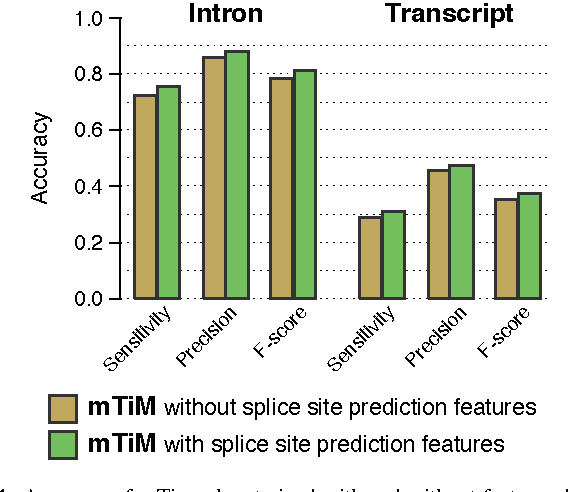
Abstract:Recent advances in high-throughput cDNA sequencing (RNA-Seq) technology have revolutionized transcriptome studies. A major motivation for RNA-Seq is to map the structure of expressed transcripts at nucleotide resolution. With accurate computational tools for transcript reconstruction, this technology may also become useful for genome (re-)annotation, which has mostly relied on de novo gene finding where gene structures are primarily inferred from the genome sequence. We developed a machine-learning method, called mTim (margin-based transcript inference method) for transcript reconstruction from RNA-Seq read alignments that is based on discriminatively trained hidden Markov support vector machines. In addition to features derived from read alignments, it utilizes characteristic genomic sequences, e.g. around splice sites, to improve transcript predictions. mTim inferred transcripts that were highly accurate and relatively robust to alignment errors in comparison to those from Cufflinks, a widely used transcript assembly method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge