Nibaran Das
An ensemble framework approach of hybrid Quantum convolutional neural networks for classification of breast cancer images
Sep 24, 2024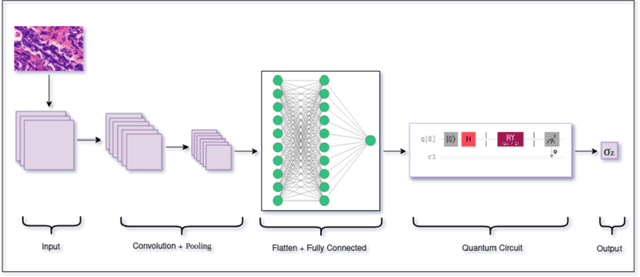
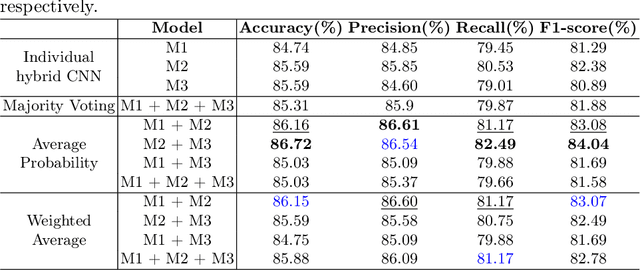

Abstract:Quantum neural networks are deemed suitable to replace classical neural networks in their ability to learn and scale up network models using quantum-exclusive phenomena like superposition and entanglement. However, in the noisy intermediate scale quantum (NISQ) era, the trainability and expressibility of quantum models are yet under investigation. Medical image classification on the other hand, pertains well to applications in deep learning, particularly, convolutional neural networks. In this paper, we carry out a study of three hybrid classical-quantum neural network architectures and combine them using standard ensembling techniques on a breast cancer histopathological dataset. The best accuracy percentage obtained by an individual model is 85.59. Whereas, on performing ensemble, we have obtained accuracy as high as 86.72%, an improvement over the individual hybrid network as well as classical neural network counterparts of the hybrid network models.
Incremental Open-set Domain Adaptation
Aug 31, 2024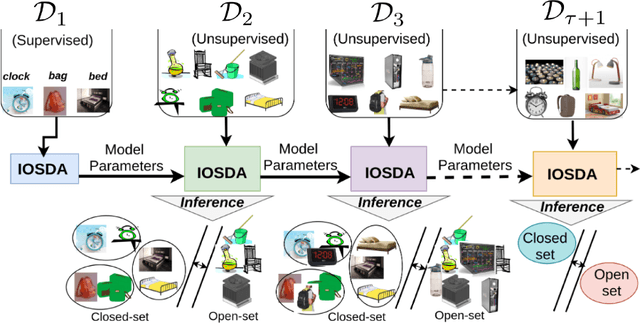
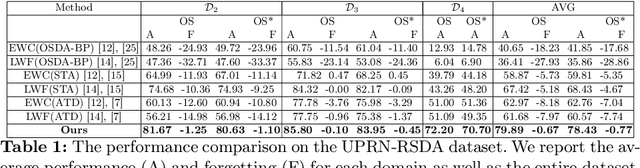
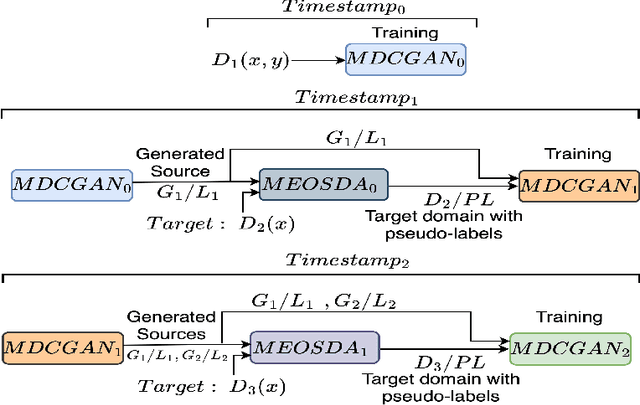
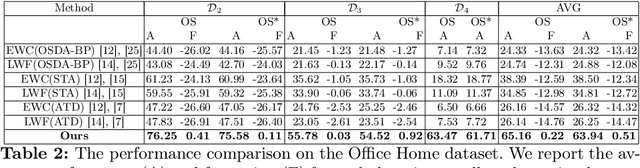
Abstract:Catastrophic forgetting makes neural network models unstable when learning visual domains consecutively. The neural network model drifts to catastrophic forgetting-induced low performance of previously learnt domains when training with new domains. We illuminate this current neural network model weakness and develop a forgetting-resistant incremental learning strategy. Here, we propose a new unsupervised incremental open-set domain adaptation (IOSDA) issue for image classification. Open-set domain adaptation adds complexity to the incremental domain adaptation issue since each target domain has more classes than the Source domain. In IOSDA, the model learns training with domain streams phase by phase in incremented time. Inference uses test data from all target domains without revealing their identities. We proposed IOSDA-Net, a two-stage learning pipeline, to solve the problem. The first module replicates prior domains from random noise using a generative framework and creates a pseudo source domain. In the second step, this pseudo source is adapted to the present target domain. We test our model on Office-Home, DomainNet, and UPRN-RSDA, a newly curated optical remote sensing dataset.
Regularizing CNNs using Confusion Penalty Based Label Smoothing for Histopathology Images
Mar 16, 2024



Abstract:Deep Learning, particularly Convolutional Neural Networks (CNN), has been successful in computer vision tasks and medical image analysis. However, modern CNNs can be overconfident, making them difficult to deploy in real-world scenarios. Researchers propose regularizing techniques, such as Label Smoothing (LS), which introduces soft labels for training data, making the classifier more regularized. LS captures disagreements or lack of confidence in the training phase, making the classifier more regularized. Although LS is quite simple and effective, traditional LS techniques utilize a weighted average between target distribution and a uniform distribution across the classes, which limits the objective of LS as well as the performance. This paper introduces a novel LS technique based on the confusion penalty, which treats model confusion for each class with more importance than others. We have performed extensive experiments with well-known CNN architectures with this technique on publicly available Colorectal Histology datasets and got satisfactory results. Also, we have compared our findings with the State-of-the-art and shown our method's efficacy with Reliability diagrams and t-distributed Stochastic Neighbor Embedding (t-SNE) plots of feature space.
COVID-CT-H-UNet: a novel COVID-19 CT segmentation network based on attention mechanism and Bi-category Hybrid loss
Mar 16, 2024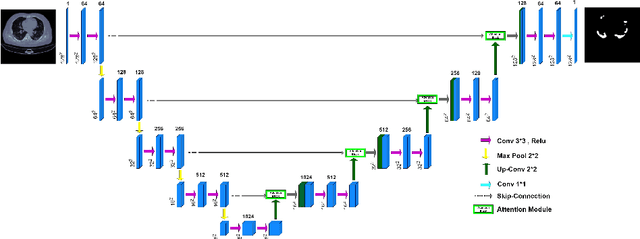

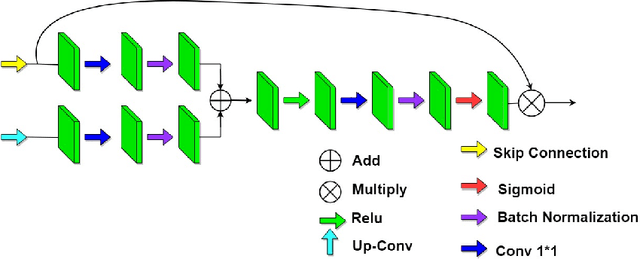

Abstract:Since 2019, the global COVID-19 outbreak has emerged as a crucial focus in healthcare research. Although RT-PCR stands as the primary method for COVID-19 detection, its extended detection time poses a significant challenge. Consequently, supplementing RT-PCR with the pathological study of COVID-19 through CT imaging has become imperative. The current segmentation approach based on TVLoss enhances the connectivity of afflicted areas. Nevertheless, it tends to misclassify normal pixels between certain adjacent diseased regions as diseased pixels. The typical Binary cross entropy(BCE) based U-shaped network only concentrates on the entire CT images without emphasizing on the affected regions, which results in hazy borders and low contrast in the projected output. In addition, the fraction of infected pixels in CT images is much less, which makes it a challenge for segmentation models to make accurate predictions. In this paper, we propose COVID-CT-H-UNet, a COVID-19 CT segmentation network to solve these problems. To recognize the unaffected pixels between neighbouring diseased regions, extra visual layer information is captured by combining the attention module on the skip connections with the proposed composite function Bi-category Hybrid Loss. The issue of hazy boundaries and poor contrast brought on by the BCE Loss in conventional techniques is resolved by utilizing the composite function Bi-category Hybrid Loss that concentrates on the pixels in the diseased area. The experiment shows when compared to the previous COVID-19 segmentation networks, the proposed COVID-CT-H-UNet's segmentation impact has greatly improved, and it may be used to identify and study clinical COVID-19.
Fuzzy Rank-based Late Fusion Technique for Cytology image Segmentation
Mar 16, 2024Abstract:Cytology image segmentation is quite challenging due to its complex cellular structure and multiple overlapping regions. On the other hand, for supervised machine learning techniques, we need a large amount of annotated data, which is costly. In recent years, late fusion techniques have given some promising performances in the field of image classification. In this paper, we have explored a fuzzy-based late fusion techniques for cytology image segmentation. This fusion rule integrates three traditional semantic segmentation models UNet, SegNet, and PSPNet. The technique is applied on two cytology image datasets, i.e., cervical cytology(HErlev) and breast cytology(JUCYT-v1) image datasets. We have achieved maximum MeanIoU score 84.27% and 83.79% on the HErlev dataset and JUCYT-v1 dataset after the proposed late fusion technique, respectively which are better than that of the traditional fusion rules such as average probability, geometric mean, Borda Count, etc. The codes of the proposed model are available on GitHub.
Could We Generate Cytology Images from Histopathology Images? An Empirical Study
Mar 16, 2024Abstract:Automation in medical imaging is quite challenging due to the unavailability of annotated datasets and the scarcity of domain experts. In recent years, deep learning techniques have solved some complex medical imaging tasks like disease classification, important object localization, segmentation, etc. However, most of the task requires a large amount of annotated data for their successful implementation. To mitigate the shortage of data, different generative models are proposed for data augmentation purposes which can boost the classification performances. For this, different synthetic medical image data generation models are developed to increase the dataset. Unpaired image-to-image translation models here shift the source domain to the target domain. In the breast malignancy identification domain, FNAC is one of the low-cost low-invasive modalities normally used by medical practitioners. But availability of public datasets in this domain is very poor. Whereas, for automation of cytology images, we need a large amount of annotated data. Therefore synthetic cytology images are generated by translating breast histopathology samples which are publicly available. In this study, we have explored traditional image-to-image transfer models like CycleGAN, and Neural Style Transfer. Further, it is observed that the generated cytology images are quite similar to real breast cytology samples by measuring FID and KID scores.
Variational Augmentation for Enhancing Historical Document Image Binarization
Nov 12, 2022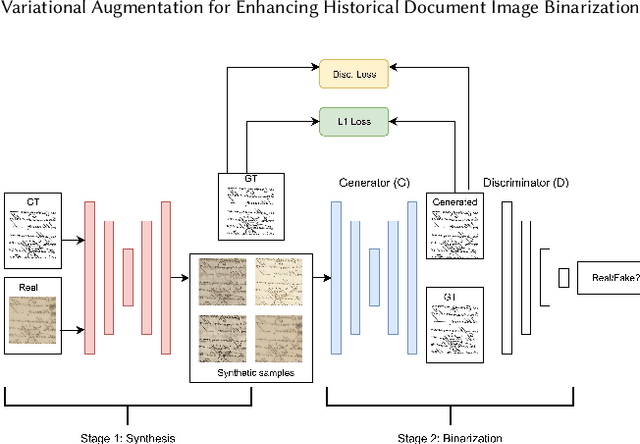
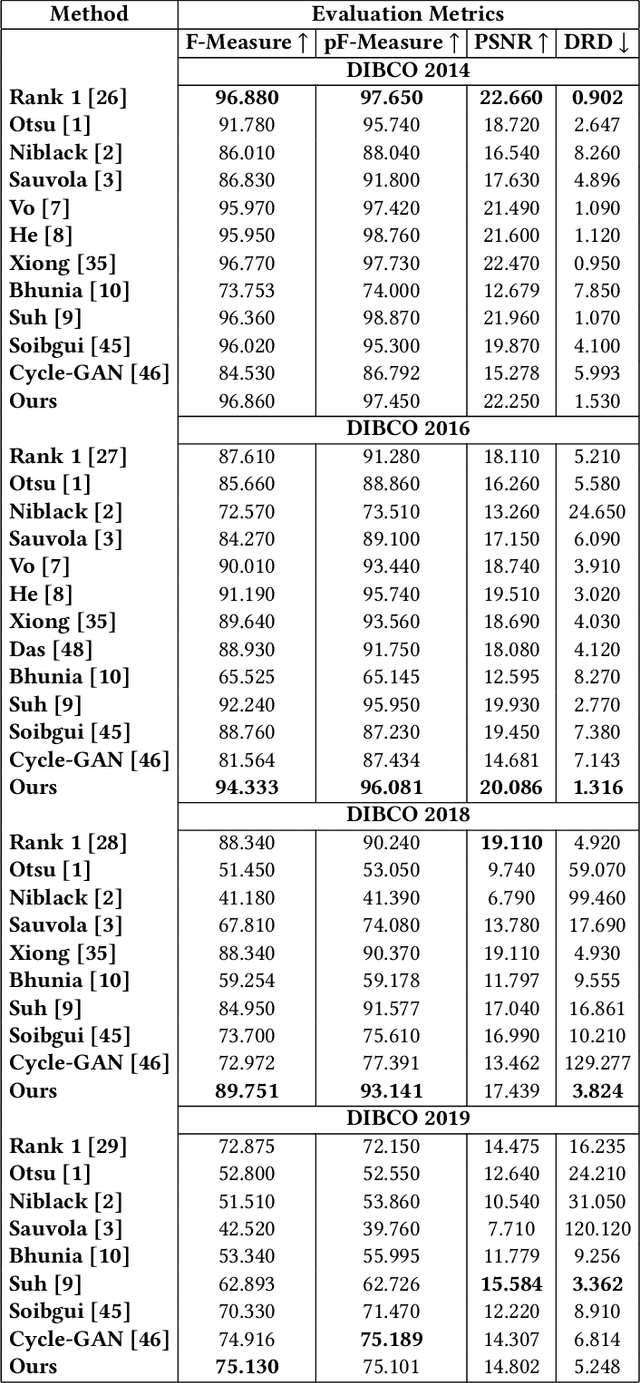
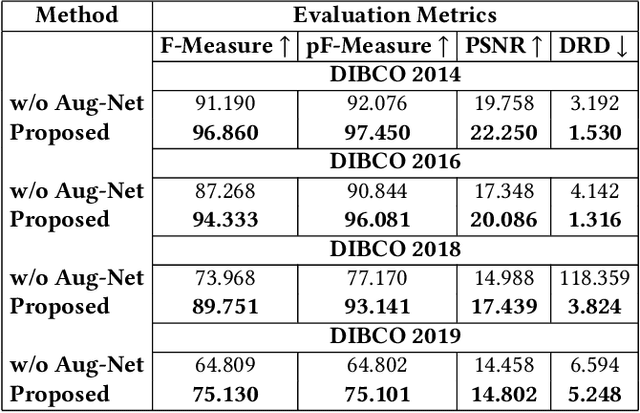
Abstract:Historical Document Image Binarization is a well-known segmentation problem in image processing. Despite ubiquity, traditional thresholding algorithms achieved limited success on severely degraded document images. With the advent of deep learning, several segmentation models were proposed that made significant progress in the field but were limited by the unavailability of large training datasets. To mitigate this problem, we have proposed a novel two-stage framework -- the first of which comprises a generator that generates degraded samples using variational inference and the second being a CNN-based binarization network that trains on the generated data. We evaluated our framework on a range of DIBCO datasets, where it achieved competitive results against previous state-of-the-art methods.
Ensemble of CNN classifiers using Sugeno Fuzzy Integral Technique for Cervical Cytology Image Classification
Aug 21, 2021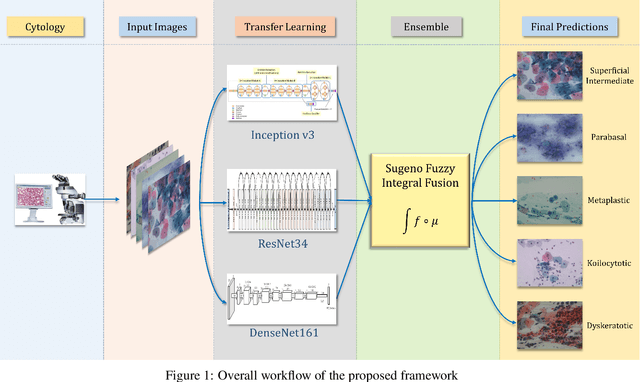
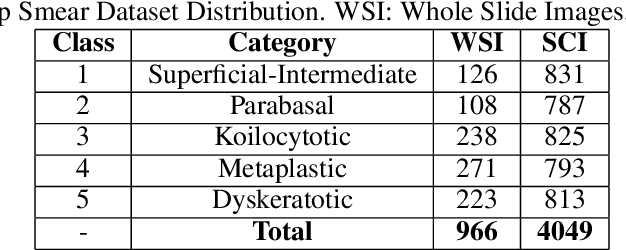
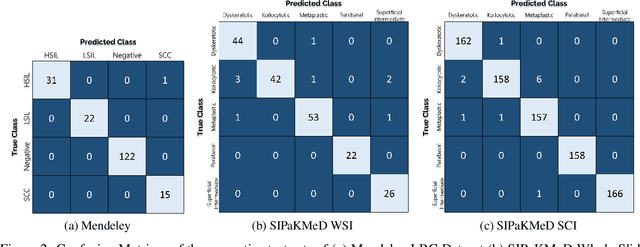
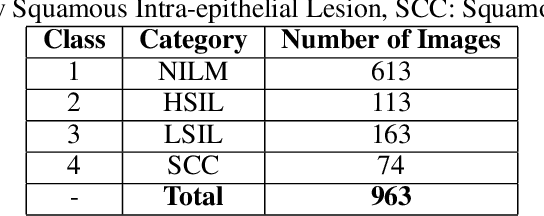
Abstract:Cervical cancer is the fourth most common category of cancer, affecting more than 500,000 women annually, owing to the slow detection procedure. Early diagnosis can help in treating and even curing cancer, but the tedious, time-consuming testing process makes it impossible to conduct population-wise screening. To aid the pathologists in efficient and reliable detection, in this paper, we propose a fully automated computer-aided diagnosis tool for classifying single-cell and slide images of cervical cancer. The main concern in developing an automatic detection tool for biomedical image classification is the low availability of publicly accessible data. Ensemble Learning is a popular approach for image classification, but simplistic approaches that leverage pre-determined weights to classifiers fail to perform satisfactorily. In this research, we use the Sugeno Fuzzy Integral to ensemble the decision scores from three popular pretrained deep learning models, namely, Inception v3, DenseNet-161 and ResNet-34. The proposed Fuzzy fusion is capable of taking into consideration the confidence scores of the classifiers for each sample, and thus adaptively changing the importance given to each classifier, capturing the complementary information supplied by each, thus leading to superior classification performance. We evaluated the proposed method on three publicly available datasets, the Mendeley Liquid Based Cytology (LBC) dataset, the SIPaKMeD Whole Slide Image (WSI) dataset, and the SIPaKMeD Single Cell Image (SCI) dataset, and the results thus yielded are promising. Analysis of the approach using GradCAM-based visual representations and statistical tests, and comparison of the method with existing and baseline models in literature justify the efficacy of the approach.
A distillation based approach for the diagnosis of diseases
Aug 07, 2021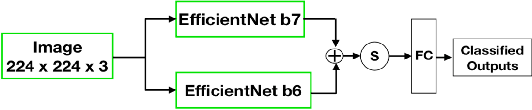
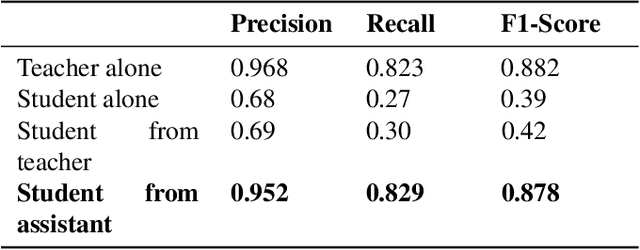
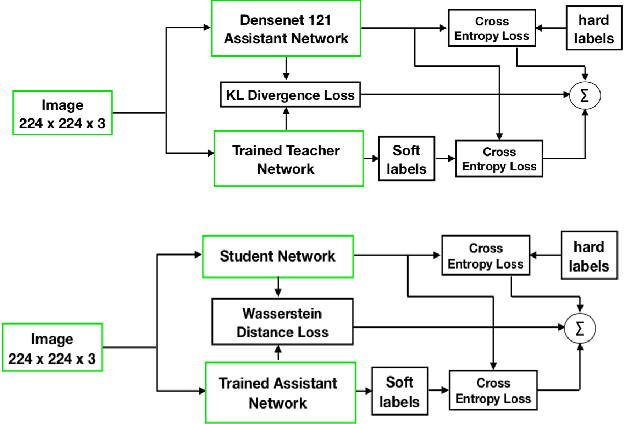
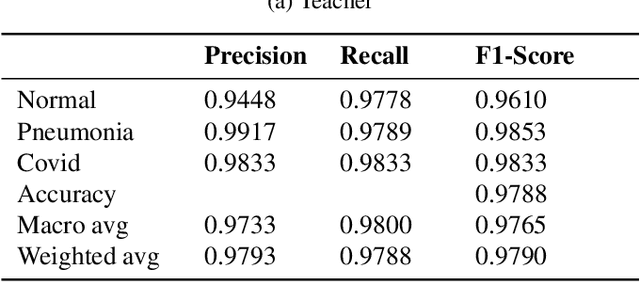
Abstract:Presently, Covid-19 is a serious threat to the world at large. Efforts are being made to reduce disease screening times and in the development of a vaccine to resist this disease, even as thousands succumb to it everyday. We propose a novel method of automated screening of diseases like Covid-19 and pneumonia from Chest X-Ray images with the help of Computer Vision. Unlike computer vision classification algorithms which come with heavy computational costs, we propose a knowledge distillation based approach which allows us to bring down the model depth, while preserving the accuracy. We make use of an augmentation of the standard distillation module with an auxiliary intermediate assistant network that aids in the continuity of the flow of information. Following this approach, we are able to build an extremely light student network, consisting of just 3 convolutional blocks without any compromise on accuracy. We thus propose a method of classification of diseases which can not only lead to faster screening, but can also operate seamlessly on low-end devices.
Cervical Cytology Classification Using PCA & GWO Enhanced Deep Features Selection
Jun 09, 2021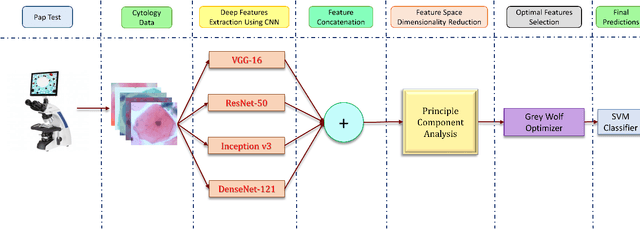
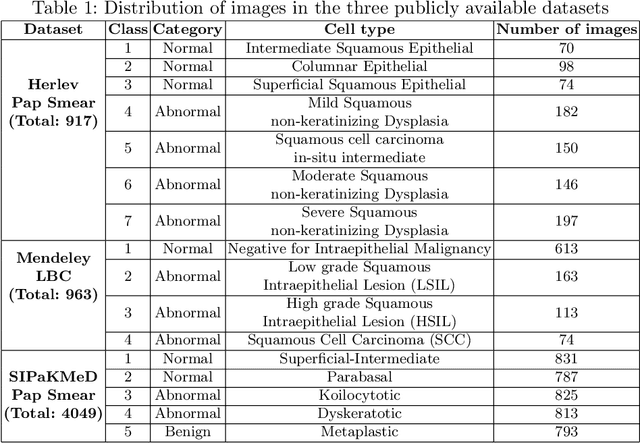
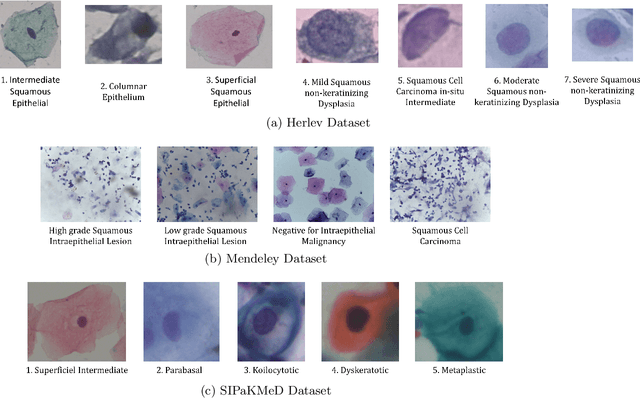
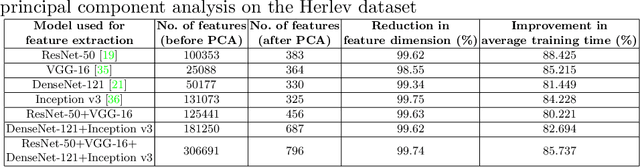
Abstract:Cervical cancer is one of the most deadly and common diseases among women worldwide. It is completely curable if diagnosed in an early stage, but the tedious and costly detection procedure makes it unviable to conduct population-wise screening. Thus, to augment the effort of the clinicians, in this paper, we propose a fully automated framework that utilizes Deep Learning and feature selection using evolutionary optimization for cytology image classification. The proposed framework extracts Deep feature from several Convolution Neural Network models and uses a two-step feature reduction approach to ensure reduction in computation cost and faster convergence. The features extracted from the CNN models form a large feature space whose dimensionality is reduced using Principal Component Analysis while preserving 99% of the variance. A non-redundant, optimal feature subset is selected from this feature space using an evolutionary optimization algorithm, the Grey Wolf Optimizer, thus improving the classification performance. Finally, the selected feature subset is used to train an SVM classifier for generating the final predictions. The proposed framework is evaluated on three publicly available benchmark datasets: Mendeley Liquid Based Cytology (4-class) dataset, Herlev Pap Smear (7-class) dataset, and the SIPaKMeD Pap Smear (5-class) dataset achieving classification accuracies of 99.47%, 98.32% and 97.87% respectively, thus justifying the reliability of the approach. The relevant codes for the proposed approach can be found in: https://github.com/DVLP-CMATERJU/Two-Step-Feature-Enhancement
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge