Sukanta Chakraborty
Fuzzy Rank-based Late Fusion Technique for Cytology image Segmentation
Mar 16, 2024Abstract:Cytology image segmentation is quite challenging due to its complex cellular structure and multiple overlapping regions. On the other hand, for supervised machine learning techniques, we need a large amount of annotated data, which is costly. In recent years, late fusion techniques have given some promising performances in the field of image classification. In this paper, we have explored a fuzzy-based late fusion techniques for cytology image segmentation. This fusion rule integrates three traditional semantic segmentation models UNet, SegNet, and PSPNet. The technique is applied on two cytology image datasets, i.e., cervical cytology(HErlev) and breast cytology(JUCYT-v1) image datasets. We have achieved maximum MeanIoU score 84.27% and 83.79% on the HErlev dataset and JUCYT-v1 dataset after the proposed late fusion technique, respectively which are better than that of the traditional fusion rules such as average probability, geometric mean, Borda Count, etc. The codes of the proposed model are available on GitHub.
Could We Generate Cytology Images from Histopathology Images? An Empirical Study
Mar 16, 2024Abstract:Automation in medical imaging is quite challenging due to the unavailability of annotated datasets and the scarcity of domain experts. In recent years, deep learning techniques have solved some complex medical imaging tasks like disease classification, important object localization, segmentation, etc. However, most of the task requires a large amount of annotated data for their successful implementation. To mitigate the shortage of data, different generative models are proposed for data augmentation purposes which can boost the classification performances. For this, different synthetic medical image data generation models are developed to increase the dataset. Unpaired image-to-image translation models here shift the source domain to the target domain. In the breast malignancy identification domain, FNAC is one of the low-cost low-invasive modalities normally used by medical practitioners. But availability of public datasets in this domain is very poor. Whereas, for automation of cytology images, we need a large amount of annotated data. Therefore synthetic cytology images are generated by translating breast histopathology samples which are publicly available. In this study, we have explored traditional image-to-image transfer models like CycleGAN, and Neural Style Transfer. Further, it is observed that the generated cytology images are quite similar to real breast cytology samples by measuring FID and KID scores.
Ensemble of CNN classifiers using Sugeno Fuzzy Integral Technique for Cervical Cytology Image Classification
Aug 21, 2021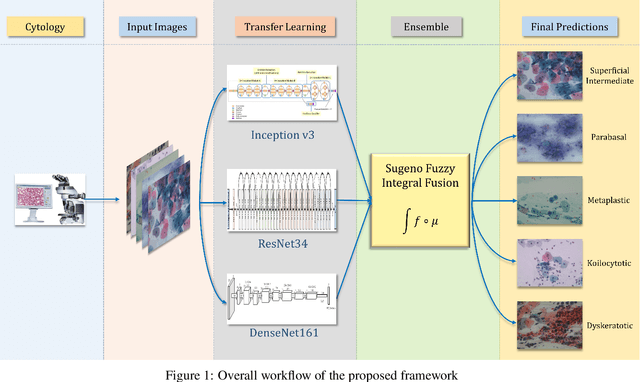
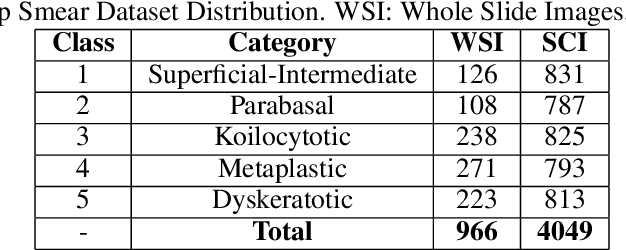
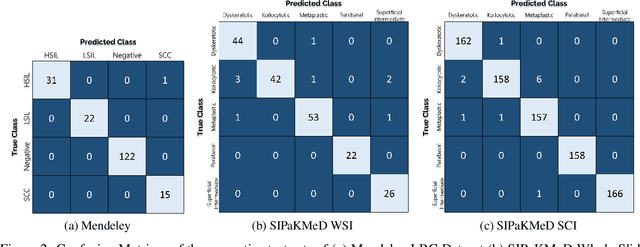
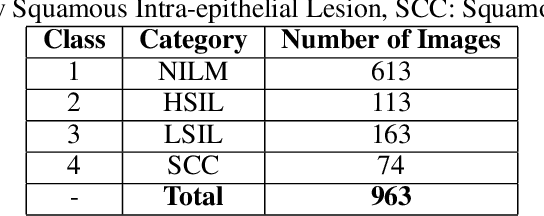
Abstract:Cervical cancer is the fourth most common category of cancer, affecting more than 500,000 women annually, owing to the slow detection procedure. Early diagnosis can help in treating and even curing cancer, but the tedious, time-consuming testing process makes it impossible to conduct population-wise screening. To aid the pathologists in efficient and reliable detection, in this paper, we propose a fully automated computer-aided diagnosis tool for classifying single-cell and slide images of cervical cancer. The main concern in developing an automatic detection tool for biomedical image classification is the low availability of publicly accessible data. Ensemble Learning is a popular approach for image classification, but simplistic approaches that leverage pre-determined weights to classifiers fail to perform satisfactorily. In this research, we use the Sugeno Fuzzy Integral to ensemble the decision scores from three popular pretrained deep learning models, namely, Inception v3, DenseNet-161 and ResNet-34. The proposed Fuzzy fusion is capable of taking into consideration the confidence scores of the classifiers for each sample, and thus adaptively changing the importance given to each classifier, capturing the complementary information supplied by each, thus leading to superior classification performance. We evaluated the proposed method on three publicly available datasets, the Mendeley Liquid Based Cytology (LBC) dataset, the SIPaKMeD Whole Slide Image (WSI) dataset, and the SIPaKMeD Single Cell Image (SCI) dataset, and the results thus yielded are promising. Analysis of the approach using GradCAM-based visual representations and statistical tests, and comparison of the method with existing and baseline models in literature justify the efficacy of the approach.
Cervical Cytology Classification Using PCA & GWO Enhanced Deep Features Selection
Jun 09, 2021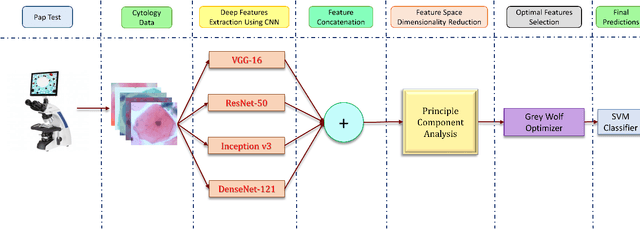
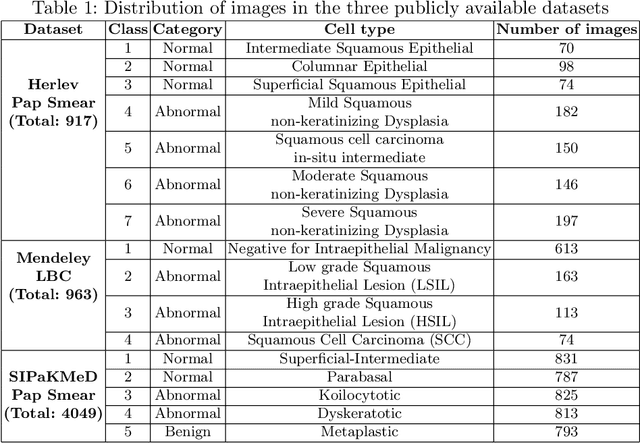
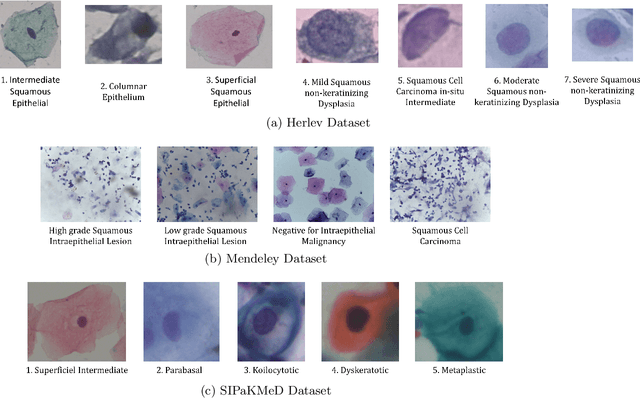
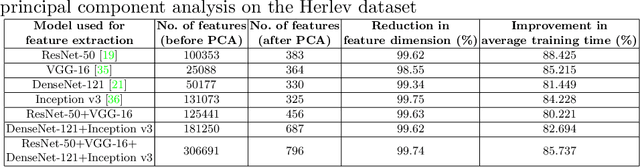
Abstract:Cervical cancer is one of the most deadly and common diseases among women worldwide. It is completely curable if diagnosed in an early stage, but the tedious and costly detection procedure makes it unviable to conduct population-wise screening. Thus, to augment the effort of the clinicians, in this paper, we propose a fully automated framework that utilizes Deep Learning and feature selection using evolutionary optimization for cytology image classification. The proposed framework extracts Deep feature from several Convolution Neural Network models and uses a two-step feature reduction approach to ensure reduction in computation cost and faster convergence. The features extracted from the CNN models form a large feature space whose dimensionality is reduced using Principal Component Analysis while preserving 99% of the variance. A non-redundant, optimal feature subset is selected from this feature space using an evolutionary optimization algorithm, the Grey Wolf Optimizer, thus improving the classification performance. Finally, the selected feature subset is used to train an SVM classifier for generating the final predictions. The proposed framework is evaluated on three publicly available benchmark datasets: Mendeley Liquid Based Cytology (4-class) dataset, Herlev Pap Smear (7-class) dataset, and the SIPaKMeD Pap Smear (5-class) dataset achieving classification accuracies of 99.47%, 98.32% and 97.87% respectively, thus justifying the reliability of the approach. The relevant codes for the proposed approach can be found in: https://github.com/DVLP-CMATERJU/Two-Step-Feature-Enhancement
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge