Myeonghyeon Kim
Anonymization for Skeleton Action Recognition
Nov 30, 2021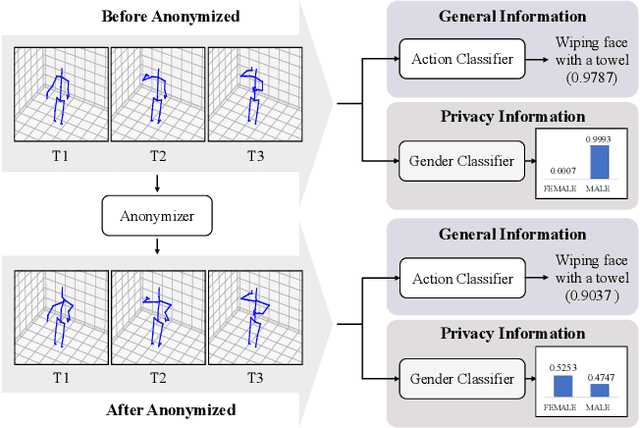
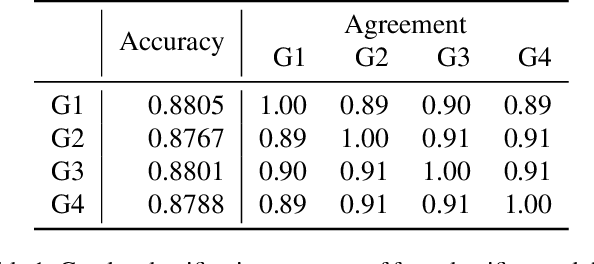
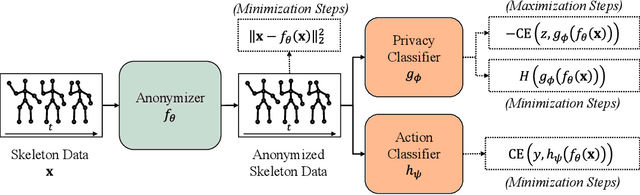
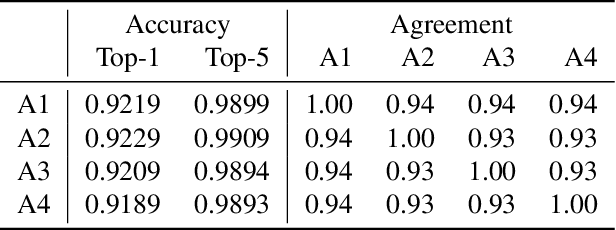
Abstract:The skeleton-based action recognition attracts practitioners and researchers due to the lightweight, compact nature of datasets. Compared with RGB-video-based action recognition, skeleton-based action recognition is a safer way to protect the privacy of subjects while having competitive recognition performance. However, due to the improvements of skeleton estimation algorithms as well as motion- and depth-sensors, more details of motion characteristics can be preserved in the skeleton dataset, leading to a potential privacy leakage from the dataset. To investigate the potential privacy leakage from the skeleton datasets, we first train a classifier to categorize sensitive private information from a trajectory of joints. Experiments show the model trained to classify gender can predict with 88% accuracy and re-identify a person with 82% accuracy. We propose two variants of anonymization algorithms to protect the potential privacy leakage from the skeleton dataset. Experimental results show that the anonymized dataset can reduce the risk of privacy leakage while having marginal effects on the action recognition performance.
2018 Robotic Scene Segmentation Challenge
Feb 03, 2020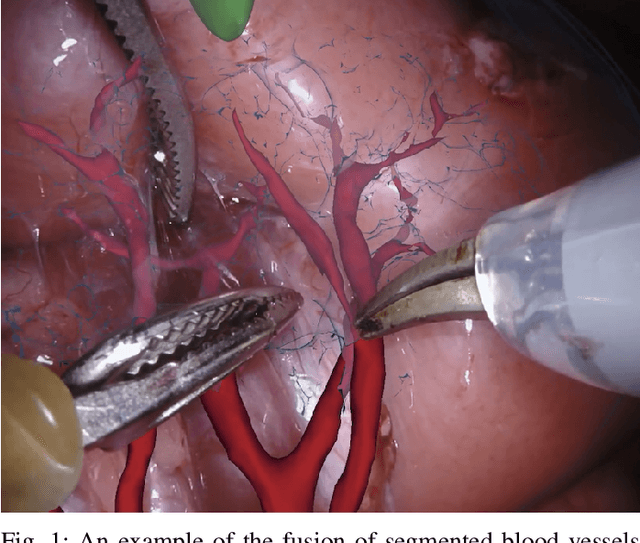
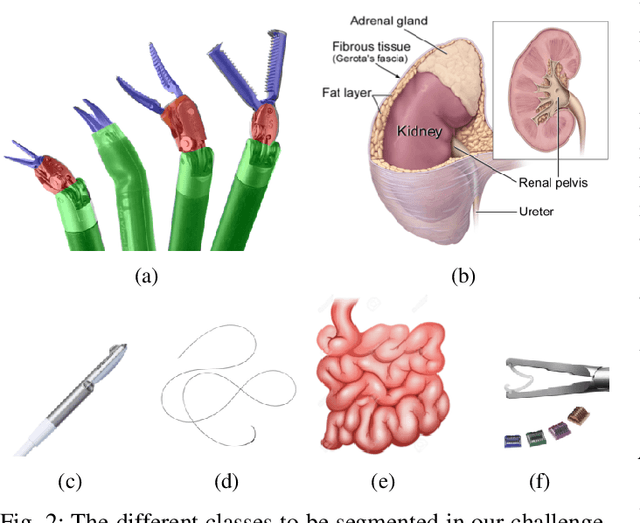
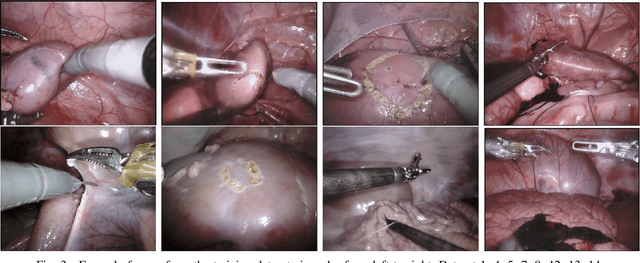
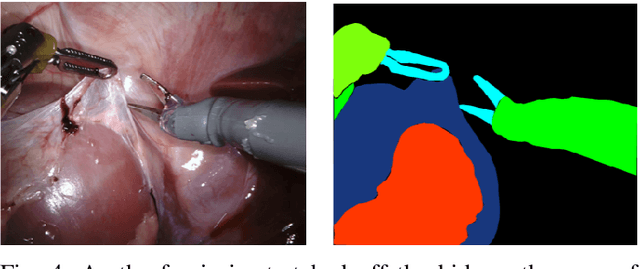
Abstract:In 2015 we began a sub-challenge at the EndoVis workshop at MICCAI in Munich using endoscope images of ex-vivo tissue with automatically generated annotations from robot forward kinematics and instrument CAD models. However, the limited background variation and simple motion rendered the dataset uninformative in learning about which techniques would be suitable for segmentation in real surgery. In 2017, at the same workshop in Quebec we introduced the robotic instrument segmentation dataset with 10 teams participating in the challenge to perform binary, articulating parts and type segmentation of da Vinci instruments. This challenge included realistic instrument motion and more complex porcine tissue as background and was widely addressed with modifications on U-Nets and other popular CNN architectures. In 2018 we added to the complexity by introducing a set of anatomical objects and medical devices to the segmented classes. To avoid over-complicating the challenge, we continued with porcine data which is dramatically simpler than human tissue due to the lack of fatty tissue occluding many organs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge