Moritz Wolf
opXRD: Open Experimental Powder X-ray Diffraction Database
Mar 07, 2025Abstract:Powder X-ray diffraction (pXRD) experiments are a cornerstone for materials structure characterization. Despite their widespread application, analyzing pXRD diffractograms still presents a significant challenge to automation and a bottleneck in high-throughput discovery in self-driving labs. Machine learning promises to resolve this bottleneck by enabling automated powder diffraction analysis. A notable difficulty in applying machine learning to this domain is the lack of sufficiently sized experimental datasets, which has constrained researchers to train primarily on simulated data. However, models trained on simulated pXRD patterns showed limited generalization to experimental patterns, particularly for low-quality experimental patterns with high noise levels and elevated backgrounds. With the Open Experimental Powder X-Ray Diffraction Database (opXRD), we provide an openly available and easily accessible dataset of labeled and unlabeled experimental powder diffractograms. Labeled opXRD data can be used to evaluate the performance of models on experimental data and unlabeled opXRD data can help improve the performance of models on experimental data, e.g. through transfer learning methods. We collected \numpatterns diffractograms, 2179 of them labeled, from a wide spectrum of materials classes. We hope this ongoing effort can guide machine learning research toward fully automated analysis of pXRD data and thus enable future self-driving materials labs.
Modular Deep Active Learning Framework for Image Annotation: A Technical Report for the Ophthalmo-AI Project
Mar 22, 2024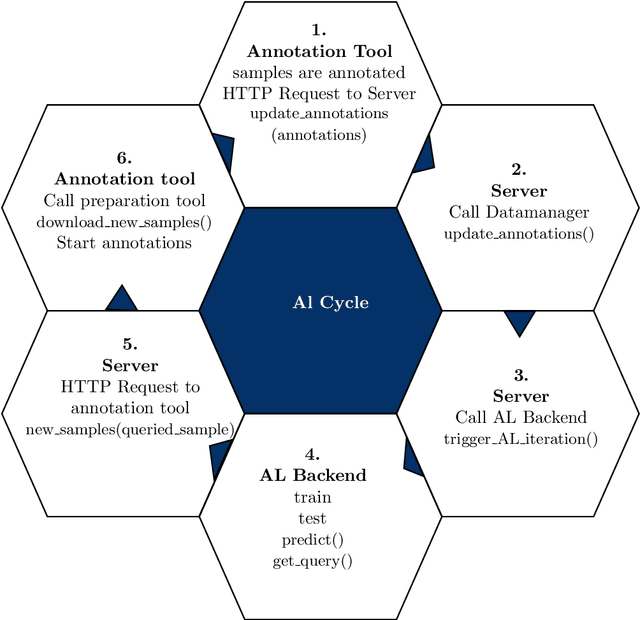
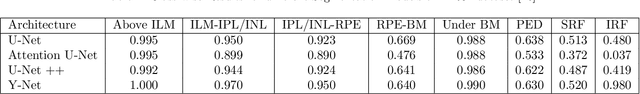
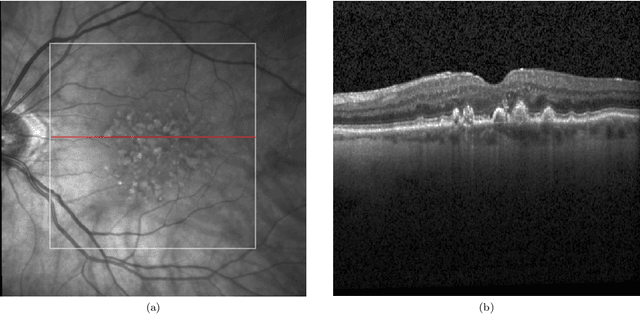
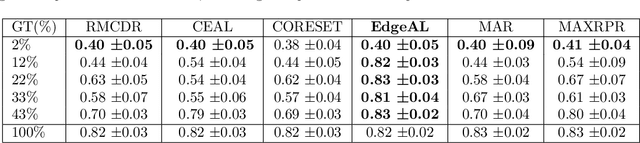
Abstract:Image annotation is one of the most essential tasks for guaranteeing proper treatment for patients and tracking progress over the course of therapy in the field of medical imaging and disease diagnosis. However, manually annotating a lot of 2D and 3D imaging data can be extremely tedious. Deep Learning (DL) based segmentation algorithms have completely transformed this process and made it possible to automate image segmentation. By accurately segmenting medical images, these algorithms can greatly minimize the time and effort necessary for manual annotation. Additionally, by incorporating Active Learning (AL) methods, these segmentation algorithms can perform far more effectively with a smaller amount of ground truth data. We introduce MedDeepCyleAL, an end-to-end framework implementing the complete AL cycle. It provides researchers with the flexibility to choose the type of deep learning model they wish to employ and includes an annotation tool that supports the classification and segmentation of medical images. The user-friendly interface allows for easy alteration of the AL and DL model settings through a configuration file, requiring no prior programming experience. While MedDeepCyleAL can be applied to any kind of image data, we have specifically applied it to ophthalmology data in this project.
HUMAN: Hierarchical Universal Modular ANnotator
Oct 02, 2020



Abstract:A lot of real-world phenomena are complex and cannot be captured by single task annotations. This causes a need for subsequent annotations, with interdependent questions and answers describing the nature of the subject at hand. Even in the case a phenomenon is easily captured by a single task, the high specialisation of most annotation tools can result in having to switch to another tool if the task only slightly changes. We introduce HUMAN, a novel web-based annotation tool that addresses the above problems by a) covering a variety of annotation tasks on both textual and image data, and b) the usage of an internal deterministic state machine, allowing the researcher to chain different annotation tasks in an interdependent manner. Further, the modular nature of the tool makes it easy to define new annotation tasks and integrate machine learning algorithms e.g., for active learning. HUMAN comes with an easy-to-use graphical user interface that simplifies the annotation task and management.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge