Hasan Md Tusfiqur Alam
CBM-RAG: Demonstrating Enhanced Interpretability in Radiology Report Generation with Multi-Agent RAG and Concept Bottleneck Models
Apr 29, 2025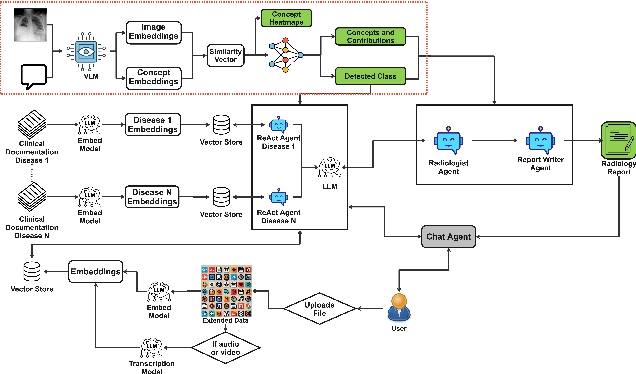
Abstract:Advancements in generative Artificial Intelligence (AI) hold great promise for automating radiology workflows, yet challenges in interpretability and reliability hinder clinical adoption. This paper presents an automated radiology report generation framework that combines Concept Bottleneck Models (CBMs) with a Multi-Agent Retrieval-Augmented Generation (RAG) system to bridge AI performance with clinical explainability. CBMs map chest X-ray features to human-understandable clinical concepts, enabling transparent disease classification. Meanwhile, the RAG system integrates multi-agent collaboration and external knowledge to produce contextually rich, evidence-based reports. Our demonstration showcases the system's ability to deliver interpretable predictions, mitigate hallucinations, and generate high-quality, tailored reports with an interactive interface addressing accuracy, trust, and usability challenges. This framework provides a pathway to improving diagnostic consistency and empowering radiologists with actionable insights.
InFL-UX: A Toolkit for Web-Based Interactive Federated Learning
Mar 06, 2025

Abstract:This paper presents InFL-UX, an interactive, proof-of-concept browser-based Federated Learning (FL) toolkit designed to integrate user contributions seamlessly into the machine learning (ML) workflow. InFL-UX enables users across multiple devices to upload datasets, define classes, and collaboratively train classification models directly in the browser using modern web technologies. Unlike traditional FL toolkits, which often focus on backend simulations, InFL-UX provides a simple user interface for researchers to explore how users interact with and contribute to FL systems in real-world, interactive settings. By prioritising usability and decentralised model training, InFL-UX bridges the gap between FL and Interactive Machine Learning (IML), empowering non-technical users to actively participate in ML classification tasks.
Enhancing Online Learning Efficiency Through Heterogeneous Resource Integration with a Multi-Agent RAG System
Feb 06, 2025


Abstract:Efficient online learning requires seamless access to diverse resources such as videos, code repositories, documentation, and general web content. This poster paper introduces early-stage work on a Multi-Agent Retrieval-Augmented Generation (RAG) System designed to enhance learning efficiency by integrating these heterogeneous resources. Using specialized agents tailored for specific resource types (e.g., YouTube tutorials, GitHub repositories, documentation websites, and search engines), the system automates the retrieval and synthesis of relevant information. By streamlining the process of finding and combining knowledge, this approach reduces manual effort and enhances the learning experience. A preliminary user study confirmed the system's strong usability and moderate-high utility, demonstrating its potential to improve the efficiency of knowledge acquisition.
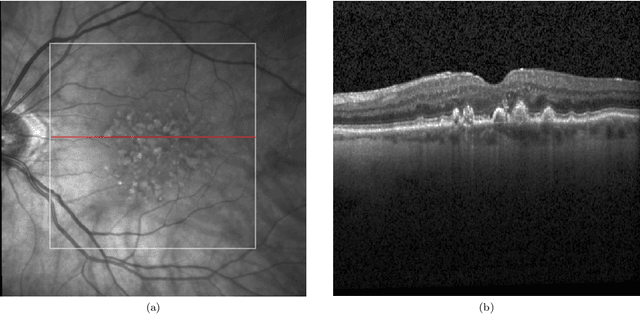
Deep Learning for Ophthalmology: The State-of-the-Art and Future Trends
Jan 07, 2025



Abstract:The emergence of artificial intelligence (AI), particularly deep learning (DL), has marked a new era in the realm of ophthalmology, offering transformative potential for the diagnosis and treatment of posterior segment eye diseases. This review explores the cutting-edge applications of DL across a range of ocular conditions, including diabetic retinopathy, glaucoma, age-related macular degeneration, and retinal vessel segmentation. We provide a comprehensive overview of foundational ML techniques and advanced DL architectures, such as CNNs, attention mechanisms, and transformer-based models, highlighting the evolving role of AI in enhancing diagnostic accuracy, optimizing treatment strategies, and improving overall patient care. Additionally, we present key challenges in integrating AI solutions into clinical practice, including ensuring data diversity, improving algorithm transparency, and effectively leveraging multimodal data. This review emphasizes AI's potential to improve disease diagnosis and enhance patient care while stressing the importance of collaborative efforts to overcome these barriers and fully harness AI's impact in advancing eye care.
Towards Interpretable Radiology Report Generation via Concept Bottlenecks using a Multi-Agentic RAG
Dec 20, 2024Abstract:Deep learning has advanced medical image classification, but interpretability challenges hinder its clinical adoption. This study enhances interpretability in Chest X-ray (CXR) classification by using concept bottleneck models (CBMs) and a multi-agent Retrieval-Augmented Generation (RAG) system for report generation. By modeling relationships between visual features and clinical concepts, we create interpretable concept vectors that guide a multi-agent RAG system to generate radiology reports, enhancing clinical relevance, explainability, and transparency. Evaluation of the generated reports using an LLM-as-a-judge confirmed the interpretability and clinical utility of our model's outputs. On the COVID-QU dataset, our model achieved 81% classification accuracy and demonstrated robust report generation performance, with five key metrics ranging between 84% and 90%. This interpretable multi-agent framework bridges the gap between high-performance AI and the explainability required for reliable AI-driven CXR analysis in clinical settings.
Modular Deep Active Learning Framework for Image Annotation: A Technical Report for the Ophthalmo-AI Project
Mar 22, 2024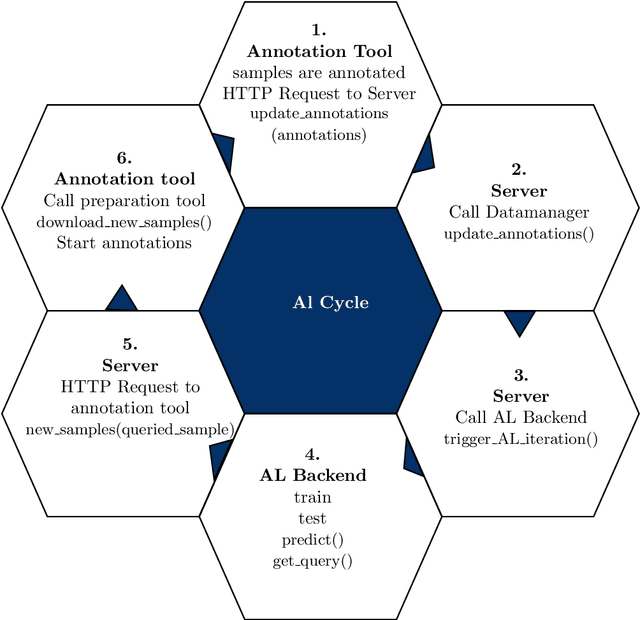
Abstract:Image annotation is one of the most essential tasks for guaranteeing proper treatment for patients and tracking progress over the course of therapy in the field of medical imaging and disease diagnosis. However, manually annotating a lot of 2D and 3D imaging data can be extremely tedious. Deep Learning (DL) based segmentation algorithms have completely transformed this process and made it possible to automate image segmentation. By accurately segmenting medical images, these algorithms can greatly minimize the time and effort necessary for manual annotation. Additionally, by incorporating Active Learning (AL) methods, these segmentation algorithms can perform far more effectively with a smaller amount of ground truth data. We introduce MedDeepCyleAL, an end-to-end framework implementing the complete AL cycle. It provides researchers with the flexibility to choose the type of deep learning model they wish to employ and includes an annotation tool that supports the classification and segmentation of medical images. The user-friendly interface allows for easy alteration of the AL and DL model settings through a configuration file, requiring no prior programming experience. While MedDeepCyleAL can be applied to any kind of image data, we have specifically applied it to ophthalmology data in this project.
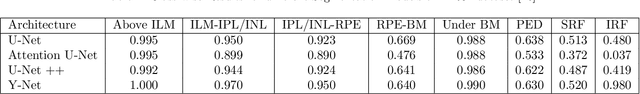
EdgeAL: An Edge Estimation Based Active Learning Approach for OCT Segmentation
Jul 25, 2023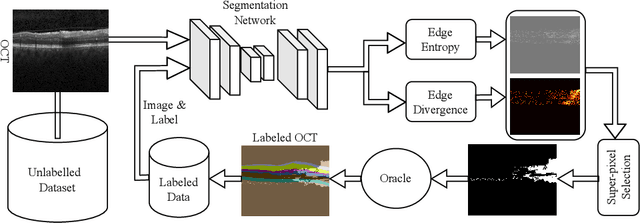
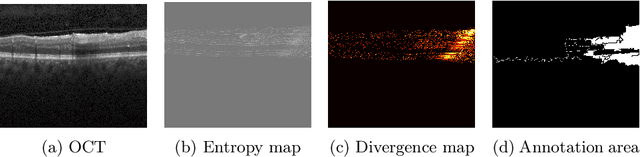

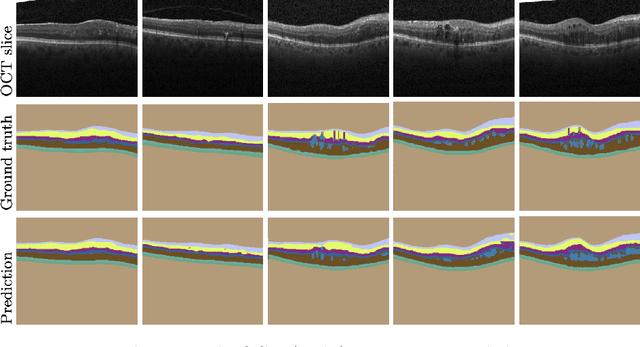
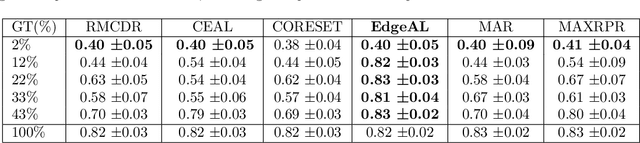
Abstract:Active learning algorithms have become increasingly popular for training models with limited data. However, selecting data for annotation remains a challenging problem due to the limited information available on unseen data. To address this issue, we propose EdgeAL, which utilizes the edge information of unseen images as {\it a priori} information for measuring uncertainty. The uncertainty is quantified by analyzing the divergence and entropy in model predictions across edges. This measure is then used to select superpixels for annotation. We demonstrate the effectiveness of EdgeAL on multi-class Optical Coherence Tomography (OCT) segmentation tasks, where we achieved a 99% dice score while reducing the annotation label cost to 12%, 2.3%, and 3%, respectively, on three publicly available datasets (Duke, AROI, and UMN). The source code is available at \url{https://github.com/Mak-Ta-Reque/EdgeAL}
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge