Hans-Jürgen Profitlich
Modular Deep Active Learning Framework for Image Annotation: A Technical Report for the Ophthalmo-AI Project
Mar 22, 2024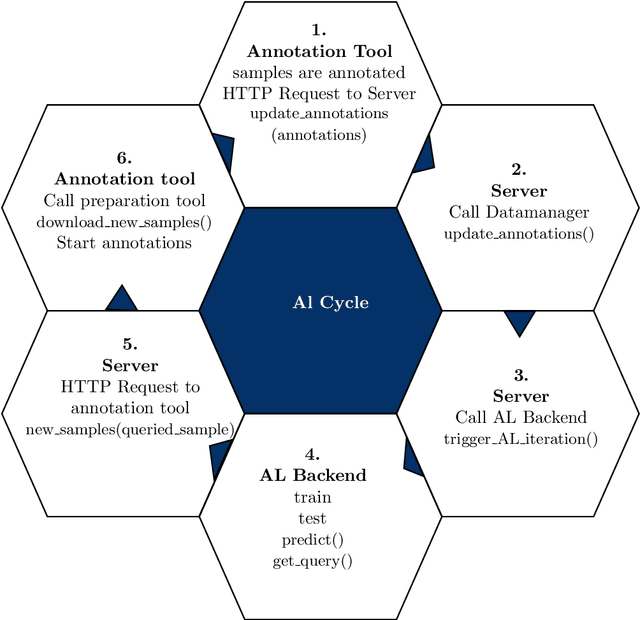
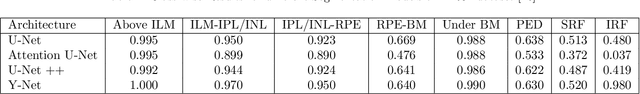
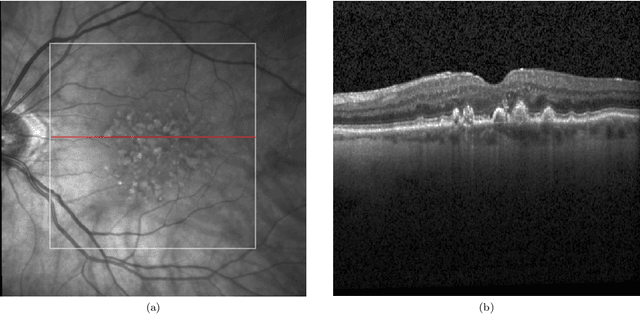
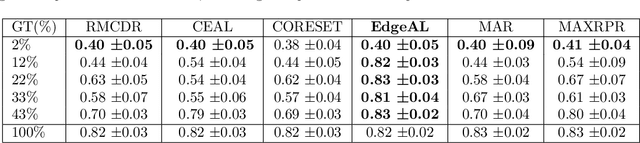
Abstract:Image annotation is one of the most essential tasks for guaranteeing proper treatment for patients and tracking progress over the course of therapy in the field of medical imaging and disease diagnosis. However, manually annotating a lot of 2D and 3D imaging data can be extremely tedious. Deep Learning (DL) based segmentation algorithms have completely transformed this process and made it possible to automate image segmentation. By accurately segmenting medical images, these algorithms can greatly minimize the time and effort necessary for manual annotation. Additionally, by incorporating Active Learning (AL) methods, these segmentation algorithms can perform far more effectively with a smaller amount of ground truth data. We introduce MedDeepCyleAL, an end-to-end framework implementing the complete AL cycle. It provides researchers with the flexibility to choose the type of deep learning model they wish to employ and includes an annotation tool that supports the classification and segmentation of medical images. The user-friendly interface allows for easy alteration of the AL and DL model settings through a configuration file, requiring no prior programming experience. While MedDeepCyleAL can be applied to any kind of image data, we have specifically applied it to ophthalmology data in this project.
A Case Study on Pros and Cons of Regular Expression Detection and Dependency Parsing for Negation Extraction from German Medical Documents. Technical Report
May 20, 2021



Abstract:We describe our work on information extraction in medical documents written in German, especially detecting negations using an architecture based on the UIMA pipeline. Based on our previous work on software modules to cover medical concepts like diagnoses, examinations, etc. we employ a version of the NegEx regular expression algorithm with a large set of triggers as a baseline. We show how a significantly smaller trigger set is sufficient to achieve similar results, in order to reduce adaptation times to new text types. We elaborate on the question whether dependency parsing (based on the Stanford CoreNLP model) is a good alternative and describe the potentials and shortcomings of both approaches.
The Skincare project, an interactive deep learning system for differential diagnosis of malignant skin lesions. Technical Report
May 19, 2020



Abstract:A shortage of dermatologists causes long wait times for patients who seek dermatologic care. In addition, the diagnostic accuracy of general practitioners has been reported to be lower than the accuracy of artificial intelligence software. This article describes the Skincare project (H2020, EIT Digital). Contributions include enabling technology for clinical decision support based on interactive machine learning (IML), a reference architecture towards a Digital European Healthcare Infrastructure (also cf. EIT MCPS), technical components for aggregating digitised patient information, and the integration of decision support technology into clinical test-bed environments. However, the main contribution is a diagnostic and decision support system in dermatology for patients and doctors, an interactive deep learning system for differential diagnosis of malignant skin lesions. In this article, we describe its functionalities and the user interfaces to facilitate machine learning from human input. The baseline deep learning system, which delivers state-of-the-art results and the potential to augment general practitioners and even dermatologists, was developed and validated using de-identified cases from a dermatology image data base (ISIC), which has about 20000 cases for development and validation, provided by board-certified dermatologists defining the reference standard for every case. ISIC allows for differential diagnosis, a ranked list of eight diagnoses, that is used to plan treatments in the common setting of diagnostic ambiguity. We give an overall description of the outcome of the Skincare project, and we focus on the steps to support communication and coordination between humans and machine in IML. This is an integral part of the development of future cognitive assistants in the medical domain, and we describe the necessary intelligent user interfaces.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge