Megan A. K. Peters
Metacognition and Uncertainty Communication in Humans and Large Language Models
Apr 18, 2025Abstract:Metacognition, the capacity to monitor and evaluate one's own knowledge and performance, is foundational to human decision-making, learning, and communication. As large language models (LLMs) become increasingly embedded in high-stakes decision contexts, it is critical to assess whether, how, and to what extent they exhibit metacognitive abilities. Here, we provide an overview of current knowledge of LLMs' metacognitive capacities, how they might be studied, and how they relate to our knowledge of metacognition in humans. We show that while humans and LLMs can sometimes appear quite aligned in their metacognitive capacities and behaviors, it is clear many differences remain. Attending to these differences is crucial not only for enhancing human-AI collaboration, but also for promoting the development of more capable and trustworthy artificial systems. Finally, we discuss how endowing future LLMs with more sensitive and more calibrated metacognition may also help them develop new capacities such as more efficient learning, self-direction, and curiosity.
Revealing higher-order neural representations with generative artificial intelligence
Mar 18, 2025Abstract:Studies often aim to reveal how neural representations encode aspects of an observer's environment, such as its contents or structure. These are ``first-order" representations (FORs), because they're ``about" the external world. A less-common target is ``higher-order" representations (HORs), which are ``about" FORs -- their contents, stability, or uncertainty. HORs of uncertainty appear critically involved in adaptive behaviors including learning under uncertainty, influencing learning rates and internal model updating based on environmental feedback. However, HORs about uncertainty are unlikely to be direct ``read-outs" of FOR characteristics, instead reflecting estimation processes which may be lossy, bias-prone, or distortive and which may also incorporate estimates of distributions of uncertainty the observer is likely to experience. While some research has targeted neural representations of ``instantaneously" estimated uncertainty, how the brain represents \textit{distributions} of expected uncertainty remains largely unexplored. Here, we propose a novel reinforcement learning (RL) based generative artificial intelligence (genAI) approach to explore neural representations of uncertainty distributions. We use existing functional magnetic resonance imaging data, where humans learned to `de-noise' their brain states to achieve target neural patterns, to train denoising diffusion genAI models with RL algorithms to learn noise distributions similar to how humans might learn to do the same. We then explore these models' learned noise-distribution HORs compared to control models trained with traditional backpropagation. Results reveal model-dependent differences in noise distribution representations -- with the RL-based model offering much higher explanatory power for human behavior -- offering an exciting path towards using genAI to explore neural noise-distribution HORs.
Bayesian temporal biclustering with applications to multi-subject neuroscience studies
Jun 24, 2024



Abstract:We consider the problem of analyzing multivariate time series collected on multiple subjects, with the goal of identifying groups of subjects exhibiting similar trends in their recorded measurements over time as well as time-varying groups of associated measurements. To this end, we propose a Bayesian model for temporal biclustering featuring nested partitions, where a time-invariant partition of subjects induces a time-varying partition of measurements. Our approach allows for data-driven determination of the number of subject and measurement clusters as well as estimation of the number and location of changepoints in measurement partitions. To efficiently perform model fitting and posterior estimation with Markov Chain Monte Carlo, we derive a blocked update of measurements' cluster-assignment sequences. We illustrate the performance of our model in two applications to functional magnetic resonance imaging data and to an electroencephalogram dataset. The results indicate that the proposed model can combine information from potentially many subjects to discover a set of interpretable, dynamic patterns. Experiments on simulated data compare the estimation performance of the proposed model against ground-truth values and other statistical methods, showing that it performs well at identifying ground-truth subject and measurement clusters even when no subject or time dependence is present.
Domain adaptation in small-scale and heterogeneous biological datasets
May 29, 2024Abstract:Machine learning techniques are steadily becoming more important in modern biology, and are used to build predictive models, discover patterns, and investigate biological problems. However, models trained on one dataset are often not generalizable to other datasets from different cohorts or laboratories, due to differences in the statistical properties of these datasets. These could stem from technical differences, such as the measurement technique used, or from relevant biological differences between the populations studied. Domain adaptation, a type of transfer learning, can alleviate this problem by aligning the statistical distributions of features and samples among different datasets so that similar models can be applied across them. However, a majority of state-of-the-art domain adaptation methods are designed to work with large-scale data, mostly text and images, while biological datasets often suffer from small sample sizes, and possess complexities such as heterogeneity of the feature space. This Review aims to synthetically discuss domain adaptation methods in the context of small-scale and highly heterogeneous biological data. We describe the benefits and challenges of domain adaptation in biological research and critically discuss some of its objectives, strengths, and weaknesses through key representative methodologies. We argue for the incorporation of domain adaptation techniques to the computational biologist's toolkit, with further development of customized approaches.
Consciousness in Artificial Intelligence: Insights from the Science of Consciousness
Aug 22, 2023
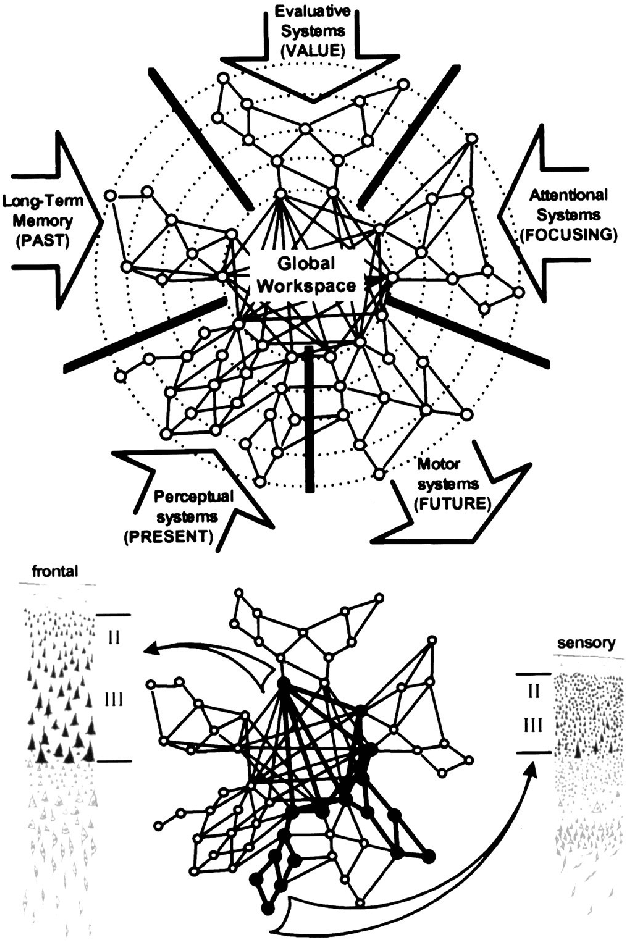

Abstract:Whether current or near-term AI systems could be conscious is a topic of scientific interest and increasing public concern. This report argues for, and exemplifies, a rigorous and empirically grounded approach to AI consciousness: assessing existing AI systems in detail, in light of our best-supported neuroscientific theories of consciousness. We survey several prominent scientific theories of consciousness, including recurrent processing theory, global workspace theory, higher-order theories, predictive processing, and attention schema theory. From these theories we derive "indicator properties" of consciousness, elucidated in computational terms that allow us to assess AI systems for these properties. We use these indicator properties to assess several recent AI systems, and we discuss how future systems might implement them. Our analysis suggests that no current AI systems are conscious, but also suggests that there are no obvious technical barriers to building AI systems which satisfy these indicators.
"Task-relevant autoencoding" enhances machine learning for human neuroscience
Aug 17, 2022



Abstract:In human neuroscience, machine learning can help reveal lower-dimensional neural representations relevant to subjects' behavior. However, state-of-the-art models typically require large datasets to train, so are prone to overfitting on human neuroimaging data that often possess few samples but many input dimensions. Here, we capitalized on the fact that the features we seek in human neuroscience are precisely those relevant to subjects' behavior. We thus developed a Task-Relevant Autoencoder via Classifier Enhancement (TRACE), and tested its ability to extract behaviorally-relevant, separable representations compared to a standard autoencoder for two severely truncated machine learning datasets. We then evaluated both models on fMRI data where subjects observed animals and objects. TRACE outperformed both the autoencoder and raw inputs nearly unilaterally, showing up to 30% increased classification accuracy and up to threefold improvement in discovering "cleaner", task-relevant representations. These results showcase TRACE's potential for a wide variety of data related to human behavior.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge