Matthieu Le
NVIDIA Nemotron Nano V2 VL
Nov 07, 2025Abstract:We introduce Nemotron Nano V2 VL, the latest model of the Nemotron vision-language series designed for strong real-world document understanding, long video comprehension, and reasoning tasks. Nemotron Nano V2 VL delivers significant improvements over our previous model, Llama-3.1-Nemotron-Nano-VL-8B, across all vision and text domains through major enhancements in model architecture, datasets, and training recipes. Nemotron Nano V2 VL builds on Nemotron Nano V2, a hybrid Mamba-Transformer LLM, and innovative token reduction techniques to achieve higher inference throughput in long document and video scenarios. We are releasing model checkpoints in BF16, FP8, and FP4 formats and sharing large parts of our datasets, recipes and training code.
Eagle 2.5: Boosting Long-Context Post-Training for Frontier Vision-Language Models
Apr 21, 2025Abstract:We introduce Eagle 2.5, a family of frontier vision-language models (VLMs) for long-context multimodal learning. Our work addresses the challenges in long video comprehension and high-resolution image understanding, introducing a generalist framework for both tasks. The proposed training framework incorporates Automatic Degrade Sampling and Image Area Preservation, two techniques that preserve contextual integrity and visual details. The framework also includes numerous efficiency optimizations in the pipeline for long-context data training. Finally, we propose Eagle-Video-110K, a novel dataset that integrates both story-level and clip-level annotations, facilitating long-video understanding. Eagle 2.5 demonstrates substantial improvements on long-context multimodal benchmarks, providing a robust solution to the limitations of existing VLMs. Notably, our best model Eagle 2.5-8B achieves 72.4% on Video-MME with 512 input frames, matching the results of top-tier commercial model such as GPT-4o and large-scale open-source models like Qwen2.5-VL-72B and InternVL2.5-78B.
Nemotron-H: A Family of Accurate and Efficient Hybrid Mamba-Transformer Models
Apr 10, 2025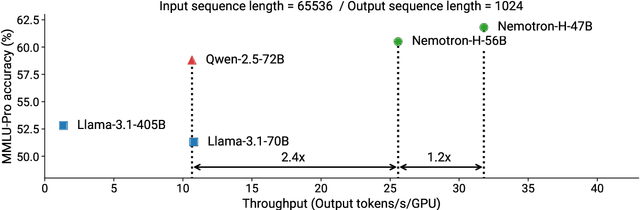

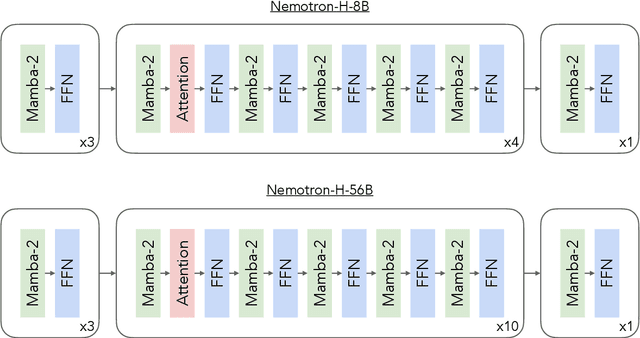
Abstract:As inference-time scaling becomes critical for enhanced reasoning capabilities, it is increasingly becoming important to build models that are efficient to infer. We introduce Nemotron-H, a family of 8B and 56B/47B hybrid Mamba-Transformer models designed to reduce inference cost for a given accuracy level. To achieve this goal, we replace the majority of self-attention layers in the common Transformer model architecture with Mamba layers that perform constant computation and require constant memory per generated token. We show that Nemotron-H models offer either better or on-par accuracy compared to other similarly-sized state-of-the-art open-sourced Transformer models (e.g., Qwen-2.5-7B/72B and Llama-3.1-8B/70B), while being up to 3$\times$ faster at inference. To further increase inference speed and reduce the memory required at inference time, we created Nemotron-H-47B-Base from the 56B model using a new compression via pruning and distillation technique called MiniPuzzle. Nemotron-H-47B-Base achieves similar accuracy to the 56B model, but is 20% faster to infer. In addition, we introduce an FP8-based training recipe and show that it can achieve on par results with BF16-based training. This recipe is used to train the 56B model. All Nemotron-H models will be released, with support in Hugging Face, NeMo, and Megatron-LM.
OMCAT: Omni Context Aware Transformer
Oct 15, 2024



Abstract:Large Language Models (LLMs) have made significant strides in text generation and comprehension, with recent advancements extending into multimodal LLMs that integrate visual and audio inputs. However, these models continue to struggle with fine-grained, cross-modal temporal understanding, particularly when correlating events across audio and video streams. We address these challenges with two key contributions: a new dataset and model, called OCTAV and OMCAT respectively. OCTAV (Omni Context and Temporal Audio Video) is a novel dataset designed to capture event transitions across audio and video. Second, OMCAT (Omni Context Aware Transformer) is a powerful model that leverages RoTE (Rotary Time Embeddings), an innovative extension of RoPE, to enhance temporal grounding and computational efficiency in time-anchored tasks. Through a robust three-stage training pipeline-feature alignment, instruction tuning, and OCTAV-specific training-OMCAT excels in cross-modal temporal understanding. Our model demonstrates state-of-the-art performance on Audio-Visual Question Answering (AVQA) tasks and the OCTAV benchmark, showcasing significant gains in temporal reasoning and cross-modal alignment, as validated through comprehensive experiments and ablation studies. Our dataset and code will be made publicly available. The link to our demo page is https://om-cat.github.io.
Computationally efficient cardiac views projection using 3D Convolutional Neural Networks
Nov 03, 2017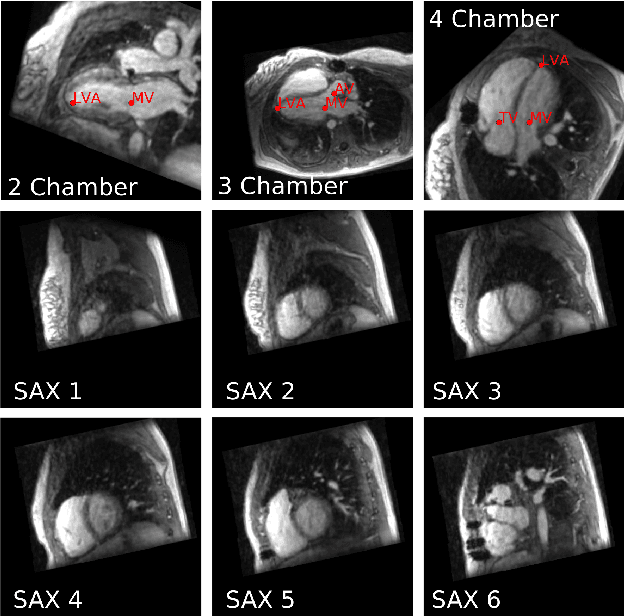
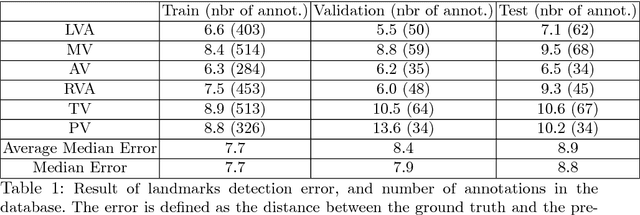
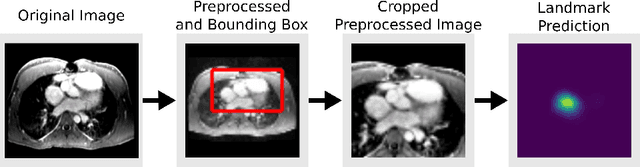
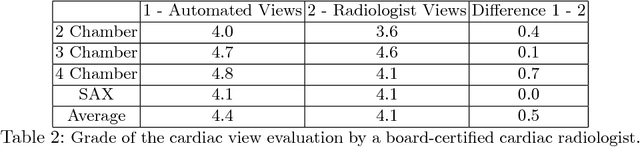
Abstract:4D Flow is an MRI sequence which allows acquisition of 3D images of the heart. The data is typically acquired volumetrically, so it must be reformatted to generate cardiac long axis and short axis views for diagnostic interpretation. These views may be generated by placing 6 landmarks: the left and right ventricle apex, and the aortic, mitral, pulmonary, and tricuspid valves. In this paper, we propose an automatic method to localize landmarks in order to compute the cardiac views. Our approach consists of first calculating a bounding box that tightly crops the heart, followed by a landmark localization step within this bounded region. Both steps are based on a 3D extension of the recently introduced ENet. We demonstrate that the long and short axis projections computed with our automated method are of equivalent quality to projections created with landmarks placed by an experienced cardiac radiologist, based on a blinded test administered to a different cardiac radiologist.
FastVentricle: Cardiac Segmentation with ENet
Apr 13, 2017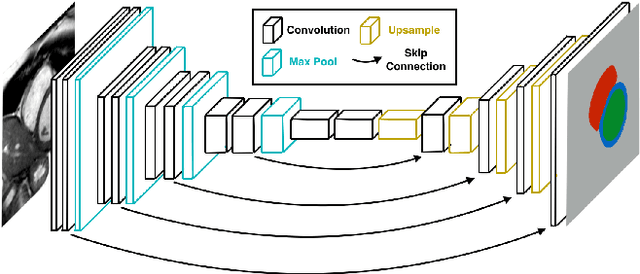
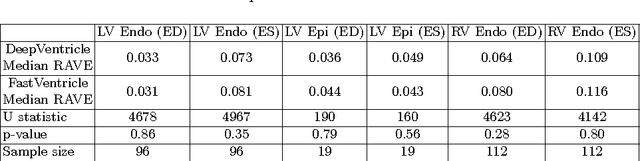
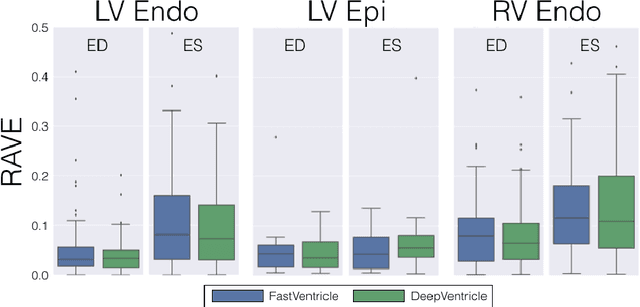
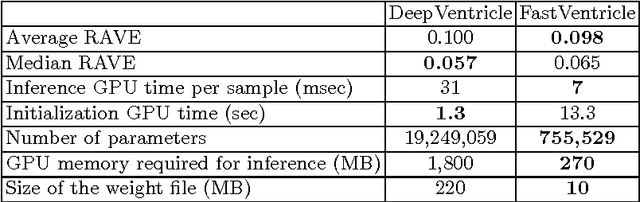
Abstract:Cardiac Magnetic Resonance (CMR) imaging is commonly used to assess cardiac structure and function. One disadvantage of CMR is that post-processing of exams is tedious. Without automation, precise assessment of cardiac function via CMR typically requires an annotator to spend tens of minutes per case manually contouring ventricular structures. Automatic contouring can lower the required time per patient by generating contour suggestions that can be lightly modified by the annotator. Fully convolutional networks (FCNs), a variant of convolutional neural networks, have been used to rapidly advance the state-of-the-art in automated segmentation, which makes FCNs a natural choice for ventricular segmentation. However, FCNs are limited by their computational cost, which increases the monetary cost and degrades the user experience of production systems. To combat this shortcoming, we have developed the FastVentricle architecture, an FCN architecture for ventricular segmentation based on the recently developed ENet architecture. FastVentricle is 4x faster and runs with 6x less memory than the previous state-of-the-art ventricular segmentation architecture while still maintaining excellent clinical accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge