Felix Lau
Natural Adversarial Objects
Nov 07, 2021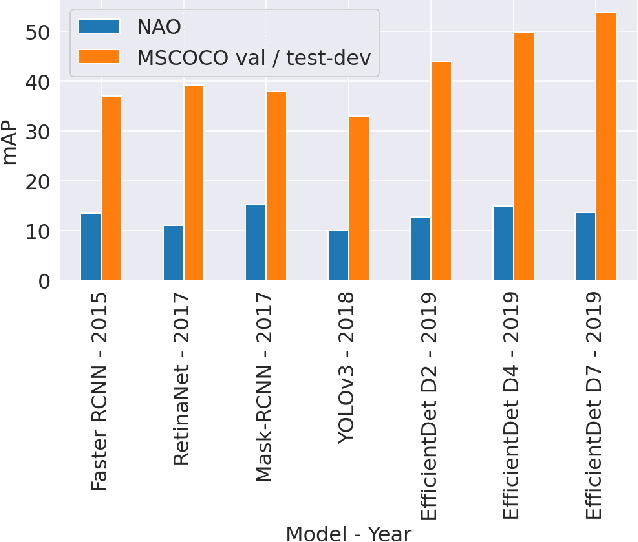


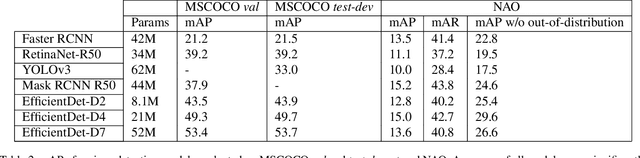
Abstract:Although state-of-the-art object detection methods have shown compelling performance, models often are not robust to adversarial attacks and out-of-distribution data. We introduce a new dataset, Natural Adversarial Objects (NAO), to evaluate the robustness of object detection models. NAO contains 7,934 images and 9,943 objects that are unmodified and representative of real-world scenarios, but cause state-of-the-art detection models to misclassify with high confidence. The mean average precision (mAP) of EfficientDet-D7 drops 74.5% when evaluated on NAO compared to the standard MSCOCO validation set. Moreover, by comparing a variety of object detection architectures, we find that better performance on MSCOCO validation set does not necessarily translate to better performance on NAO, suggesting that robustness cannot be simply achieved by training a more accurate model. We further investigate why examples in NAO are difficult to detect and classify. Experiments of shuffling image patches reveal that models are overly sensitive to local texture. Additionally, using integrated gradients and background replacement, we find that the detection model is reliant on pixel information within the bounding box, and insensitive to the background context when predicting class labels. NAO can be downloaded at https://drive.google.com/drive/folders/15P8sOWoJku6SSEiHLEts86ORfytGezi8.
On The State of Data In Computer Vision: Human Annotations Remain Indispensable for Developing Deep Learning Models
Jul 31, 2021Abstract:High-quality labeled datasets play a crucial role in fueling the development of machine learning (ML), and in particular the development of deep learning (DL). However, since the emergence of the ImageNet dataset and the AlexNet model in 2012, the size of new open-source labeled vision datasets has remained roughly constant. Consequently, only a minority of publications in the computer vision community tackle supervised learning on datasets that are orders of magnitude larger than Imagenet. In this paper, we survey computer vision research domains that study the effects of such large datasets on model performance across different vision tasks. We summarize the community's current understanding of those effects, and highlight some open questions related to training with massive datasets. In particular, we tackle: (a) The largest datasets currently used in computer vision research and the interesting takeaways from training on such datasets; (b) The effectiveness of pre-training on large datasets; (c) Recent advancements and hurdles facing synthetic datasets; (d) An overview of double descent and sample non-monotonicity phenomena; and finally, (e) A brief discussion of lifelong/continual learning and how it fares compared to learning from huge labeled datasets in an offline setting. Overall, our findings are that research on optimization for deep learning focuses on perfecting the training routine and thus making DL models less data hungry, while research on synthetic datasets aims to offset the cost of data labeling. However, for the time being, acquiring non-synthetic labeled data remains indispensable to boost performance.
Evaluating Deep Neural Networks Trained on Clinical Images in Dermatology with the Fitzpatrick 17k Dataset
Apr 20, 2021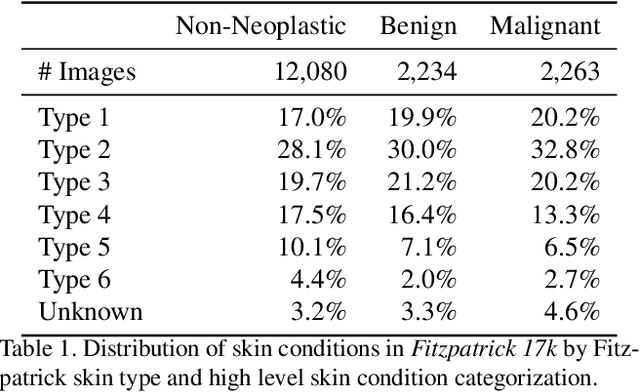
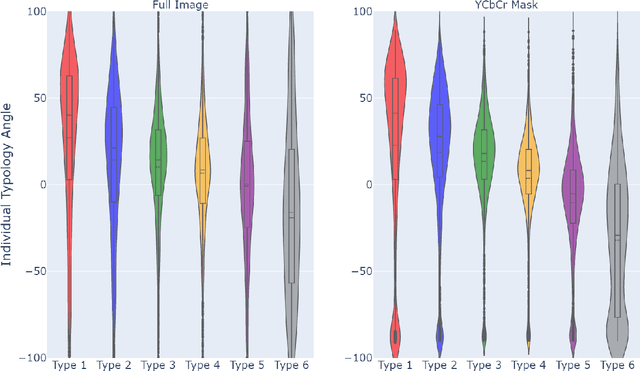
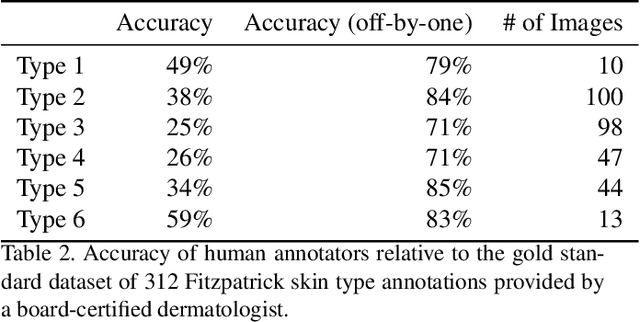
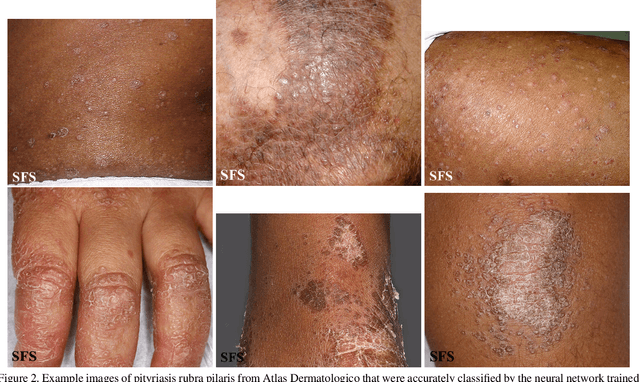
Abstract:How does the accuracy of deep neural network models trained to classify clinical images of skin conditions vary across skin color? While recent studies demonstrate computer vision models can serve as a useful decision support tool in healthcare and provide dermatologist-level classification on a number of specific tasks, darker skin is underrepresented in the data. Most publicly available data sets do not include Fitzpatrick skin type labels. We annotate 16,577 clinical images sourced from two dermatology atlases with Fitzpatrick skin type labels and open-source these annotations. Based on these labels, we find that there are significantly more images of light skin types than dark skin types in this dataset. We train a deep neural network model to classify 114 skin conditions and find that the model is most accurate on skin types similar to those it was trained on. In addition, we evaluate how an algorithmic approach to identifying skin tones, individual typology angle, compares with Fitzpatrick skin type labels annotated by a team of human labelers.
ScarGAN: Chained Generative Adversarial Networks to Simulate Pathological Tissue on Cardiovascular MR Scans
Aug 14, 2018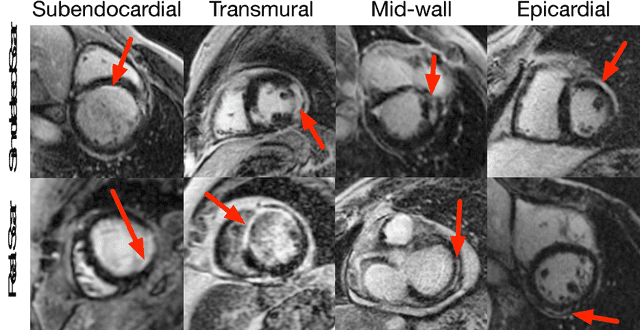
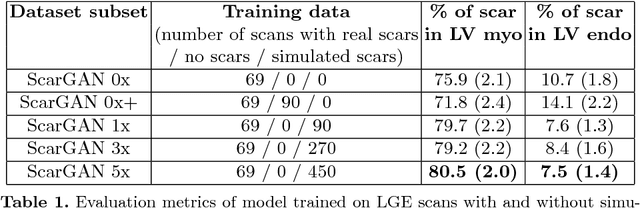
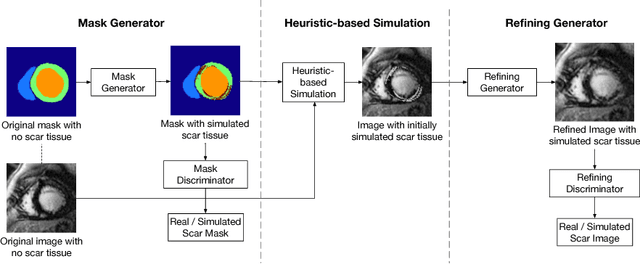
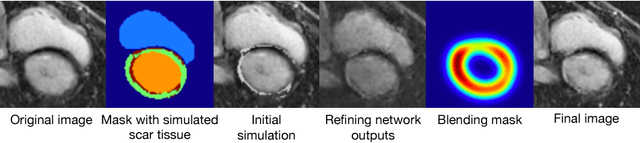
Abstract:Medical images with specific pathologies are scarce, but a large amount of data is usually required for a deep convolutional neural network (DCNN) to achieve good accuracy. We consider the problem of segmenting the left ventricular (LV) myocardium on late gadolinium enhancement (LGE) cardiovascular magnetic resonance (CMR) scans of which only some of the scans have scar tissue. We propose ScarGAN to simulate scar tissue on healthy myocardium using chained generative adversarial networks (GAN). Our novel approach factorizes the simulation process into 3 steps: 1) a mask generator to simulate the shape of the scar tissue; 2) a domain-specific heuristic to produce the initial simulated scar tissue from the simulated shape; 3) a refining generator to add details to the simulated scar tissue. Unlike other approaches that generate samples from scratch, we simulate scar tissue on normal scans resulting in highly realistic samples. We show that experienced radiologists are unable to distinguish between real and simulated scar tissue. Training a U-Net with additional scans with scar tissue simulated by ScarGAN increases the percentage of scar pixels correctly included in LV myocardium prediction from 75.9% to 80.5%.
Computationally efficient cardiac views projection using 3D Convolutional Neural Networks
Nov 03, 2017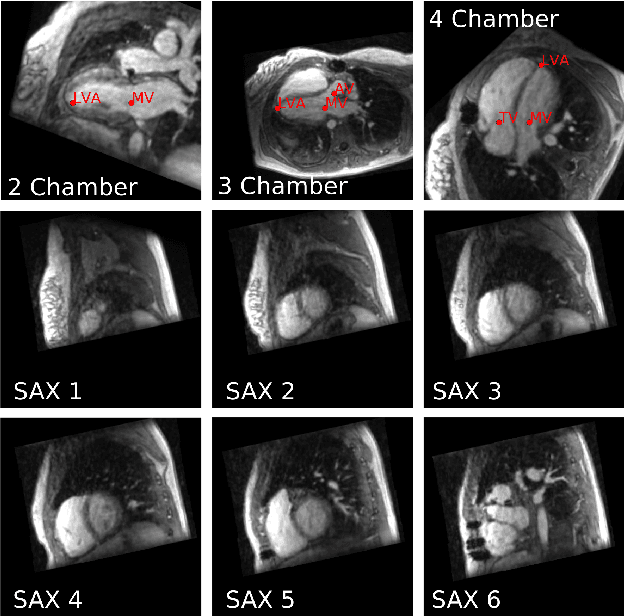
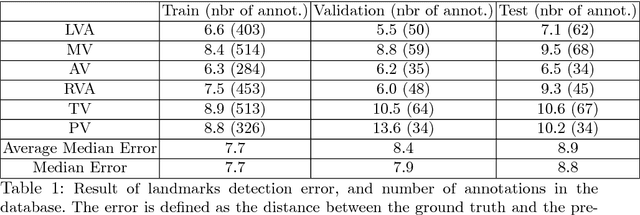
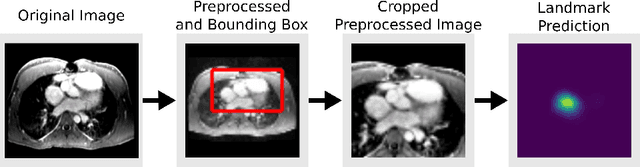
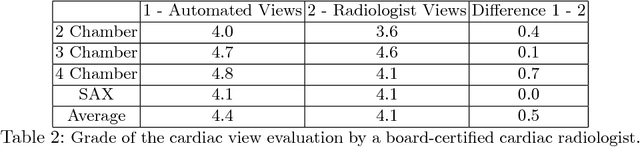
Abstract:4D Flow is an MRI sequence which allows acquisition of 3D images of the heart. The data is typically acquired volumetrically, so it must be reformatted to generate cardiac long axis and short axis views for diagnostic interpretation. These views may be generated by placing 6 landmarks: the left and right ventricle apex, and the aortic, mitral, pulmonary, and tricuspid valves. In this paper, we propose an automatic method to localize landmarks in order to compute the cardiac views. Our approach consists of first calculating a bounding box that tightly crops the heart, followed by a landmark localization step within this bounded region. Both steps are based on a 3D extension of the recently introduced ENet. We demonstrate that the long and short axis projections computed with our automated method are of equivalent quality to projections created with landmarks placed by an experienced cardiac radiologist, based on a blinded test administered to a different cardiac radiologist.
FastVentricle: Cardiac Segmentation with ENet
Apr 13, 2017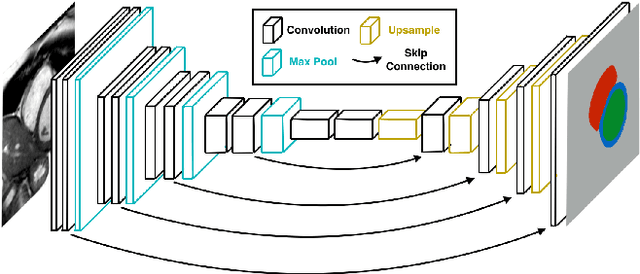
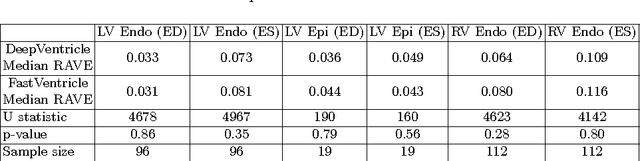
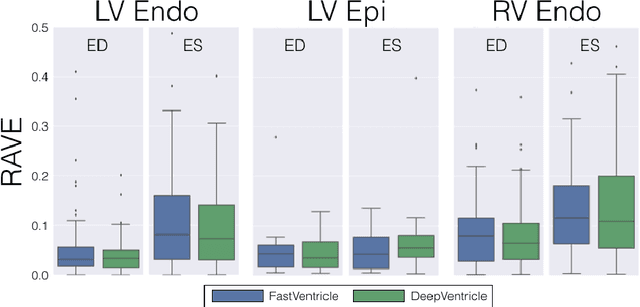
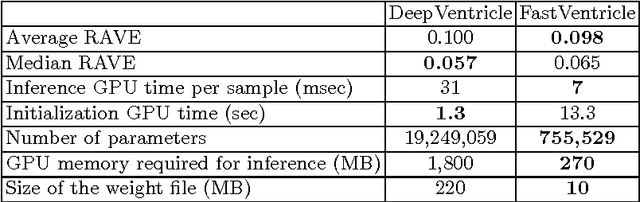
Abstract:Cardiac Magnetic Resonance (CMR) imaging is commonly used to assess cardiac structure and function. One disadvantage of CMR is that post-processing of exams is tedious. Without automation, precise assessment of cardiac function via CMR typically requires an annotator to spend tens of minutes per case manually contouring ventricular structures. Automatic contouring can lower the required time per patient by generating contour suggestions that can be lightly modified by the annotator. Fully convolutional networks (FCNs), a variant of convolutional neural networks, have been used to rapidly advance the state-of-the-art in automated segmentation, which makes FCNs a natural choice for ventricular segmentation. However, FCNs are limited by their computational cost, which increases the monetary cost and degrades the user experience of production systems. To combat this shortcoming, we have developed the FastVentricle architecture, an FCN architecture for ventricular segmentation based on the recently developed ENet architecture. FastVentricle is 4x faster and runs with 6x less memory than the previous state-of-the-art ventricular segmentation architecture while still maintaining excellent clinical accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge