Matthias Grimm
Motion-Aware Robotic 3D Ultrasound
Jul 13, 2021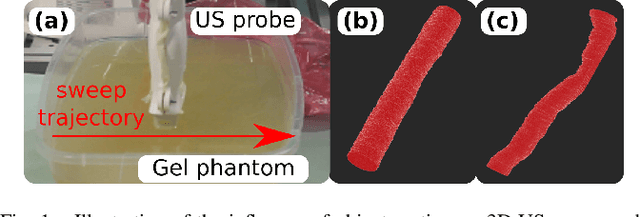
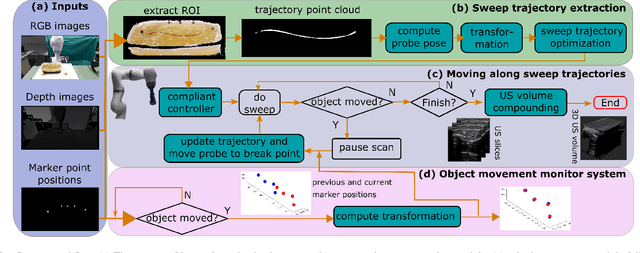
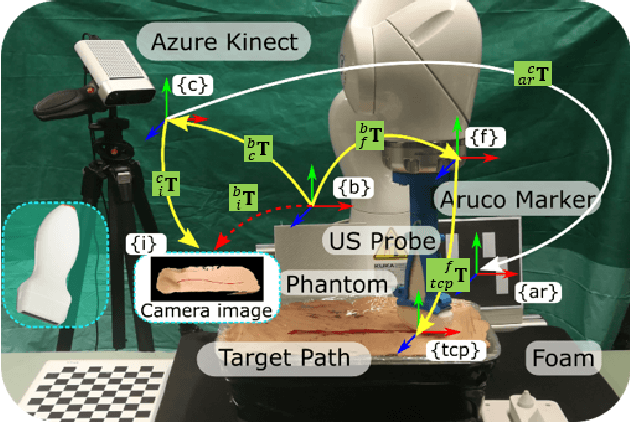
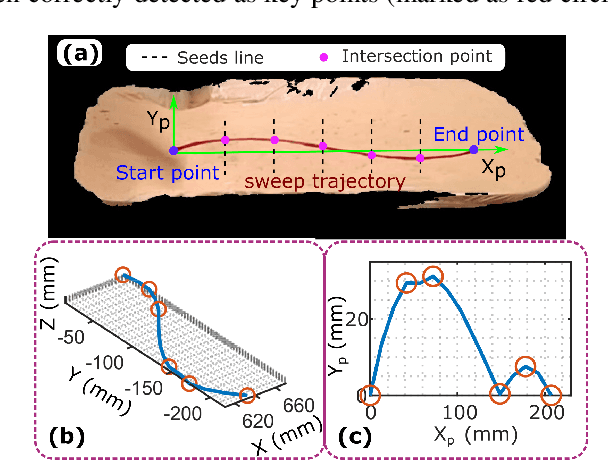
Abstract:Robotic three-dimensional (3D) ultrasound (US) imaging has been employed to overcome the drawbacks of traditional US examinations, such as high inter-operator variability and lack of repeatability. However, object movement remains a challenge as unexpected motion decreases the quality of the 3D compounding. Furthermore, attempted adjustment of objects, e.g., adjusting limbs to display the entire limb artery tree, is not allowed for conventional robotic US systems. To address this challenge, we propose a vision-based robotic US system that can monitor the object's motion and automatically update the sweep trajectory to provide 3D compounded images of the target anatomy seamlessly. To achieve these functions, a depth camera is employed to extract the manually planned sweep trajectory after which the normal direction of the object is estimated using the extracted 3D trajectory. Subsequently, to monitor the movement and further compensate for this motion to accurately follow the trajectory, the position of firmly attached passive markers is tracked in real-time. Finally, a step-wise compounding was performed. The experiments on a gel phantom demonstrate that the system can resume a sweep when the object is not stationary during scanning.
Pose-dependent weights and Domain Randomization for fully automatic X-ray to CT Registration
Nov 14, 2020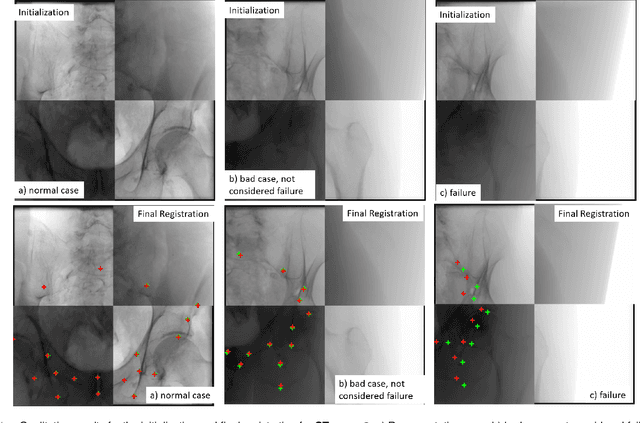
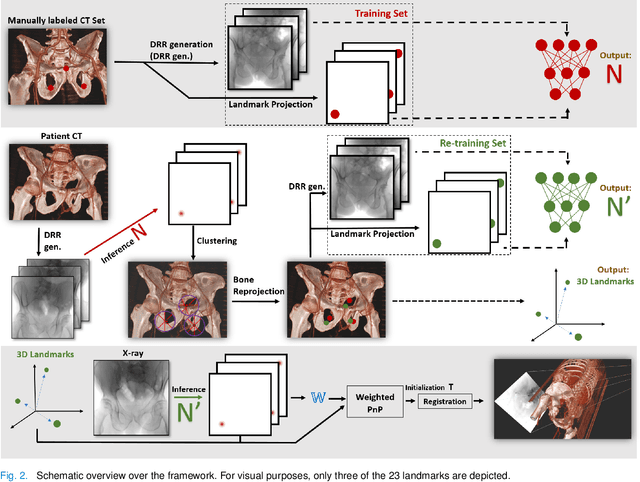
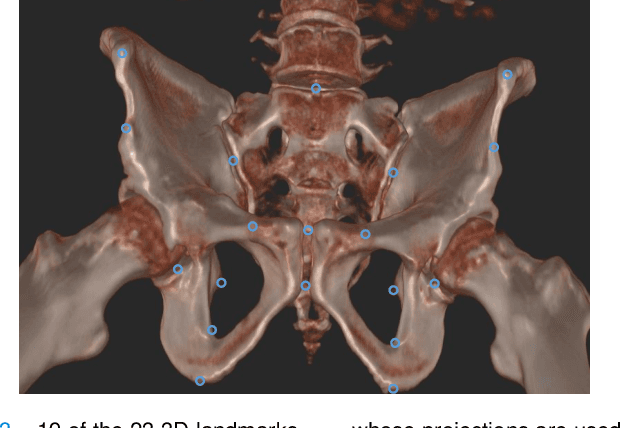
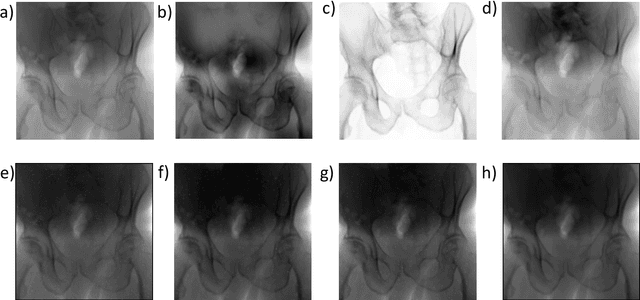
Abstract:Fully automatic X-ray to CT registration requires a solid initialization to provide an initial alignment within the capture range of existing intensity-based registrations. This work adresses that need by providing a novel automatic initialization, which enables end to end registration. First, a neural network is trained once to detect a set of anatomical landmarks on simulated X-rays. A domain randomization scheme is proposed to enable the network to overcome the challenge of being trained purely on simulated data and run inference on real Xrays. Then, for each patient CT, a patient-specific landmark extraction scheme is used. It is based on backprojecting and clustering the previously trained networks predictions on a set of simulated X-rays. Next, the network is retrained to detect the new landmarks. Finally the combination of network and 3D landmark locations is used to compute the initialization using a perspective-n-point algorithm. During the computation of the pose, a weighting scheme is introduced to incorporate the confidence of the network in detecting the landmarks. The algorithm is evaluated on the pelvis using both real and simulated x-rays. The mean (+-standard deviation) target registration error in millimetres is 4.1 +- 4.3 for simulated X-rays with a success rate of 92% and 4.2 +- 3.9 for real X-rays with a success rate of 86.8%, where a success is defined as a translation error of less than 30mm.
Autonomous Robotic Screening of Tubular Structures based only on Real-Time Ultrasound Imaging Feedback
Oct 30, 2020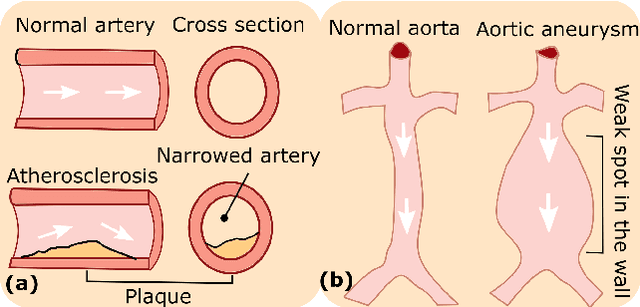
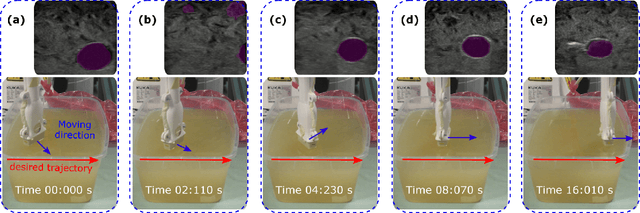
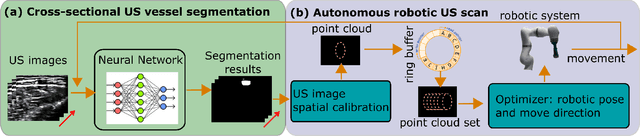
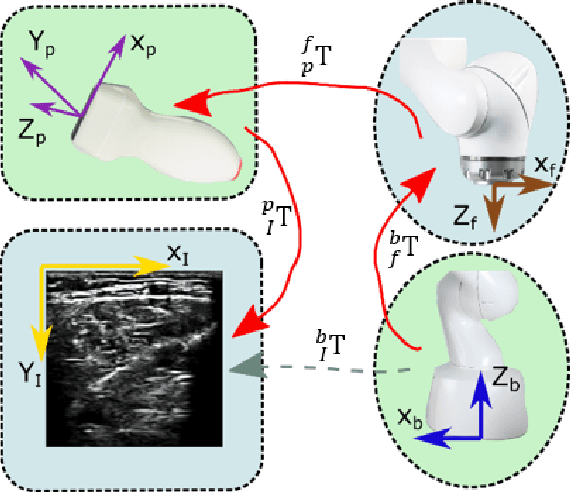
Abstract:Ultrasound (US) imaging is widely employed for diagnosis and staging of peripheral vascular diseases (PVD), mainly due to its high availability and the fact it does not emit radiation. However, high inter-operator variability and a lack of repeatability of US image acquisition hinder the implementation of extensive screening programs. To address this challenge, we propose an end-to-end workflow for automatic robotic US screening of tubular structures using only the real-time US imaging feedback. We first train a U-Net for real-time segmentation of the vascular structure from cross-sectional US images. Then, we represent the detected vascular structure as a 3D point cloud and use it to estimate the longitudinal axis of the target tubular structure and its mean radius by solving a constrained non-linear optimization problem. Iterating the previous processes, the US probe is automatically aligned to the orientation normal to the target tubular tissue and adjusted online to center the tracked tissue based on the spatial calibration. The real-time segmentation result is evaluated both on a phantom and in-vivo on brachial arteries of volunteers. In addition, the whole process is validated both in simulation and physical phantoms. The mean absolute radius error and orientation error ($\pm$ SD) in the simulation are $1.16\pm0.1~mm$ and $2.7\pm3.3^{\circ}$, respectively. On a gel phantom, these errors are $1.95\pm2.02~mm$ and $3.3\pm2.4^{\circ}$. This shows that the method is able to automatically screen tubular tissues with an optimal probe orientation (i.e. normal to the vessel) and at the same to accurately estimate the mean radius, both in real-time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge