Mateusz Buda
Deep Learning for Classification of Thyroid Nodules on Ultrasound: Validation on an Independent Dataset
Jul 27, 2022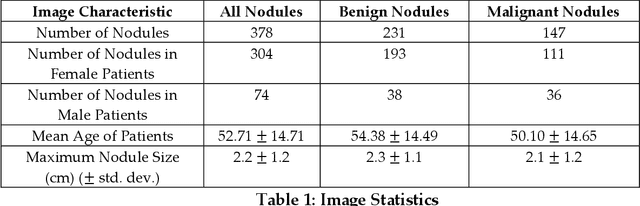
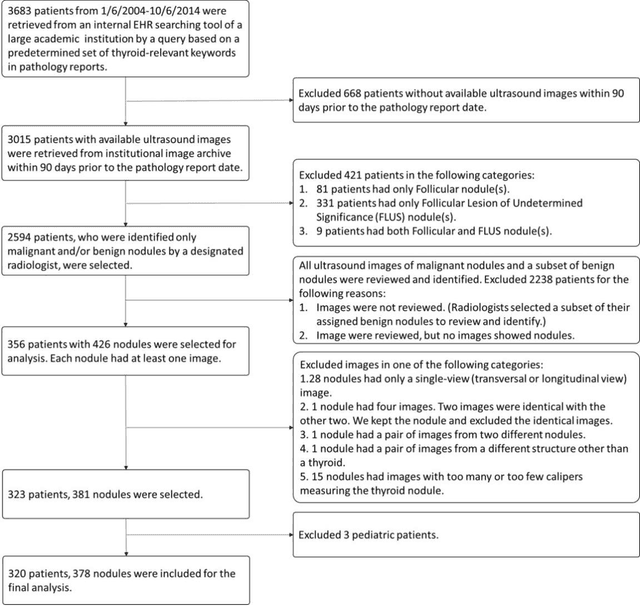
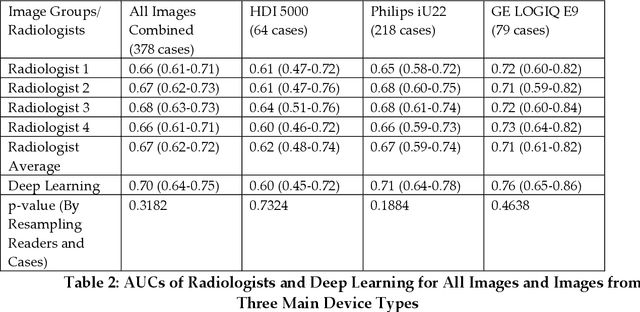
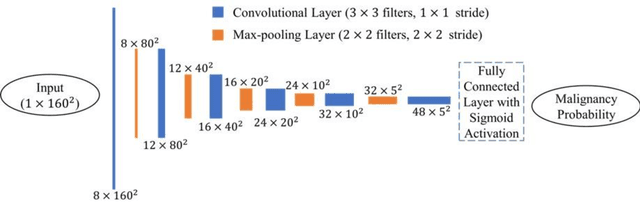
Abstract:Objectives: The purpose is to apply a previously validated deep learning algorithm to a new thyroid nodule ultrasound image dataset and compare its performances with radiologists. Methods: Prior study presented an algorithm which is able to detect thyroid nodules and then make malignancy classifications with two ultrasound images. A multi-task deep convolutional neural network was trained from 1278 nodules and originally tested with 99 separate nodules. The results were comparable with that of radiologists. The algorithm was further tested with 378 nodules imaged with ultrasound machines from different manufacturers and product types than the training cases. Four experienced radiologists were requested to evaluate the nodules for comparison with deep learning. Results: The Area Under Curve (AUC) of the deep learning algorithm and four radiologists were calculated with parametric, binormal estimation. For the deep learning algorithm, the AUC was 0.70 (95% CI: 0.64 - 0.75). The AUC of radiologists were 0.66 (95% CI: 0.61 - 0.71), 0.67 (95% CI:0.62 - 0.73), 0.68 (95% CI: 0.63 - 0.73), and 0.66 (95%CI: 0.61 - 0.71). Conclusion: In the new testing dataset, the deep learning algorithm achieved similar performances with all four radiologists.
Detection of masses and architectural distortions in digital breast tomosynthesis: a publicly available dataset of 5,060 patients and a deep learning model
Nov 13, 2020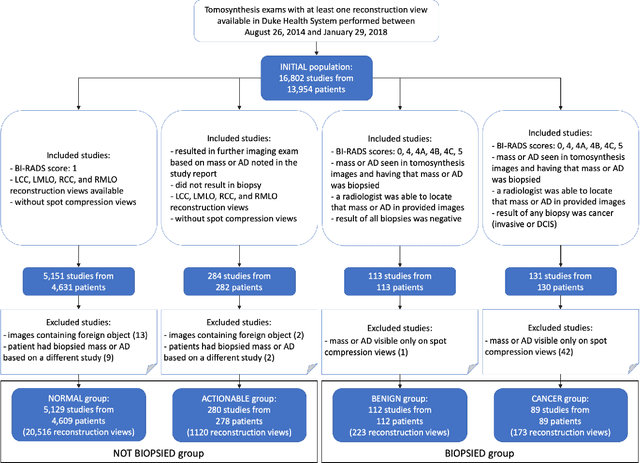
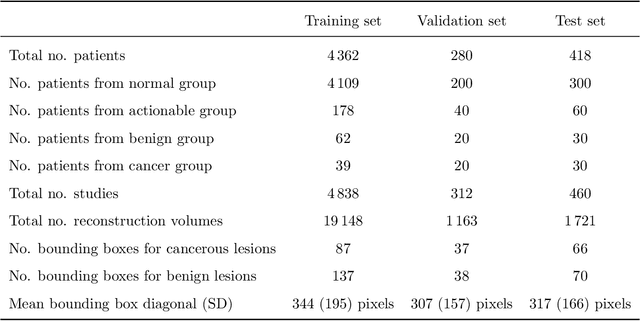
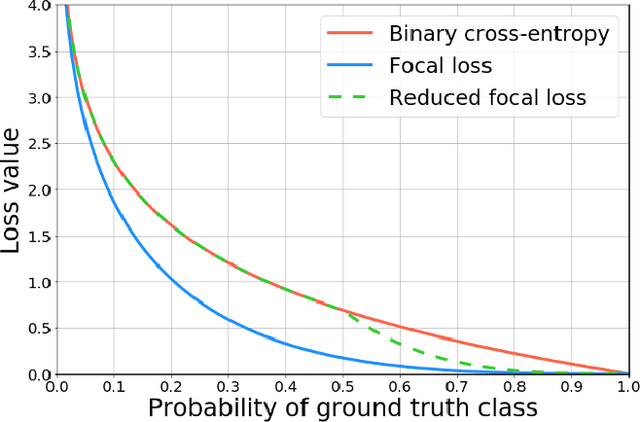
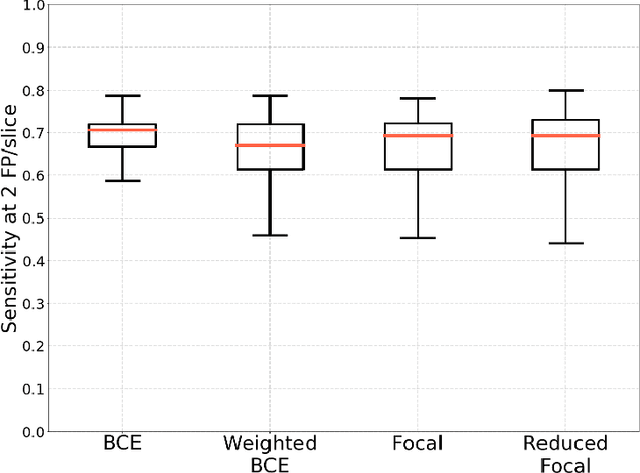
Abstract:Breast cancer screening is one of the most common radiological tasks with over 39 million exams performed each year. While breast cancer screening has been one of the most studied medical imaging applications of artificial intelligence, the development and evaluation of the algorithms are hindered due to the lack of well-annotated large-scale publicly available datasets. This is particularly an issue for digital breast tomosynthesis (DBT) which is a relatively new breast cancer screening modality. We have curated and made publicly available a large-scale dataset of digital breast tomosynthesis images. It contains 22,032 reconstructed DBT volumes belonging to 5,610 studies from 5,060 patients. This included four groups: (1) 5,129 normal studies, (2) 280 studies where additional imaging was needed but no biopsy was performed, (3) 112 benign biopsied studies, and (4) 89 studies with cancer. Our dataset included masses and architectural distortions which were annotated by two experienced radiologists. Additionally, we developed a single-phase deep learning detection model and tested it using our dataset to serve as a baseline for future research. Our model reached a sensitivity of 65% at 2 false positives per breast. Our large, diverse, and highly-curated dataset will facilitate development and evaluation of AI algorithms for breast cancer screening through providing data for training as well as common set of cases for model validation. The performance of the model developed in our study shows that the task remains challenging and will serve as a baseline for future model development.
Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm
Jun 09, 2019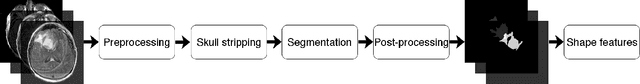
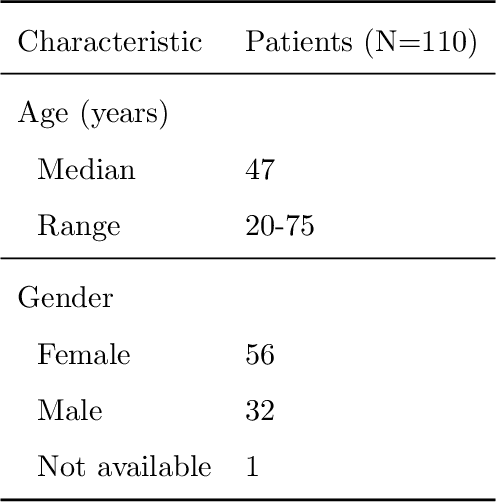
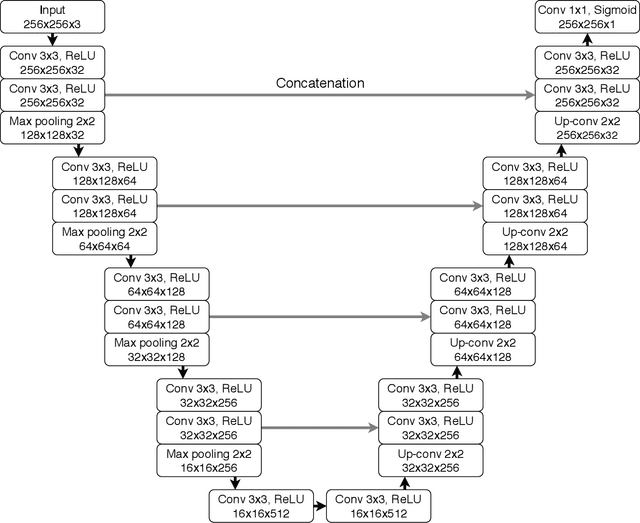
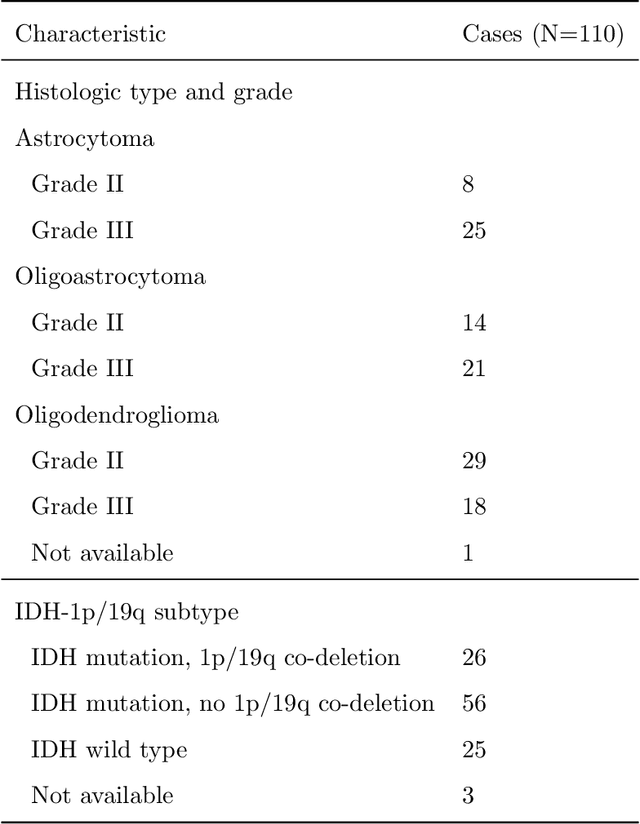
Abstract:Recent analysis identified distinct genomic subtypes of lower-grade glioma tumors which are associated with shape features. In this study, we propose a fully automatic way to quantify tumor imaging characteristics using deep learning-based segmentation and test whether these characteristics are predictive of tumor genomic subtypes. We used preoperative imaging and genomic data of 110 patients from 5 institutions with lower-grade gliomas from The Cancer Genome Atlas. Based on automatic deep learning segmentations, we extracted three features which quantify two-dimensional and three-dimensional characteristics of the tumors. Genomic data for the analyzed cohort of patients consisted of previously identified genomic clusters based on IDH mutation and 1p/19q co-deletion, DNA methylation, gene expression, DNA copy number, and microRNA expression. To analyze the relationship between the imaging features and genomic clusters, we conducted the Fisher exact test for 10 hypotheses for each pair of imaging feature and genomic subtype. To account for multiple hypothesis testing, we applied a Bonferroni correction. P-values lower than 0.005 were considered statistically significant. We found the strongest association between RNASeq clusters and the bounding ellipsoid volume ratio ($p<0.0002$) and between RNASeq clusters and margin fluctuation ($p<0.005$). In addition, we identified associations between bounding ellipsoid volume ratio and all tested molecular subtypes ($p<0.02$) as well as between angular standard deviation and RNASeq cluster ($p<0.02$). In terms of automatic tumor segmentation that was used to generate the quantitative image characteristics, our deep learning algorithm achieved a mean Dice coefficient of 82% which is comparable to human performance.
A systematic study of the class imbalance problem in convolutional neural networks
Oct 13, 2018



Abstract:In this study, we systematically investigate the impact of class imbalance on classification performance of convolutional neural networks (CNNs) and compare frequently used methods to address the issue. Class imbalance is a common problem that has been comprehensively studied in classical machine learning, yet very limited systematic research is available in the context of deep learning. In our study, we use three benchmark datasets of increasing complexity, MNIST, CIFAR-10 and ImageNet, to investigate the effects of imbalance on classification and perform an extensive comparison of several methods to address the issue: oversampling, undersampling, two-phase training, and thresholding that compensates for prior class probabilities. Our main evaluation metric is area under the receiver operating characteristic curve (ROC AUC) adjusted to multi-class tasks since overall accuracy metric is associated with notable difficulties in the context of imbalanced data. Based on results from our experiments we conclude that (i) the effect of class imbalance on classification performance is detrimental; (ii) the method of addressing class imbalance that emerged as dominant in almost all analyzed scenarios was oversampling; (iii) oversampling should be applied to the level that completely eliminates the imbalance, whereas the optimal undersampling ratio depends on the extent of imbalance; (iv) as opposed to some classical machine learning models, oversampling does not cause overfitting of CNNs; (v) thresholding should be applied to compensate for prior class probabilities when overall number of properly classified cases is of interest.
Deep learning in radiology: an overview of the concepts and a survey of the state of the art
Feb 10, 2018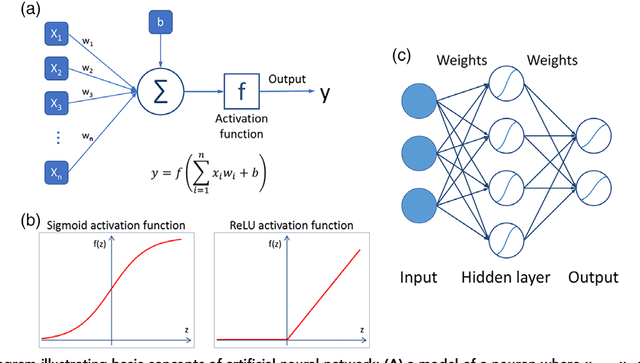
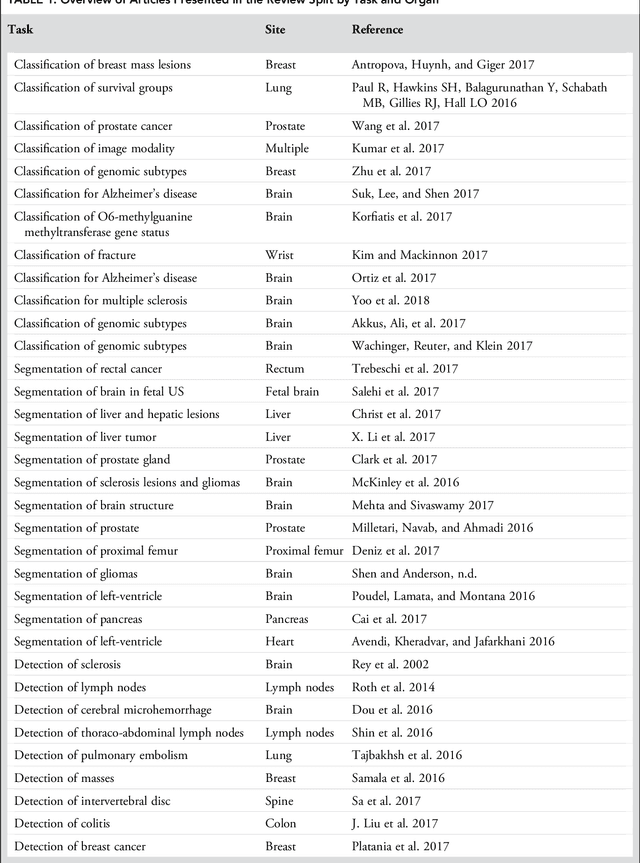
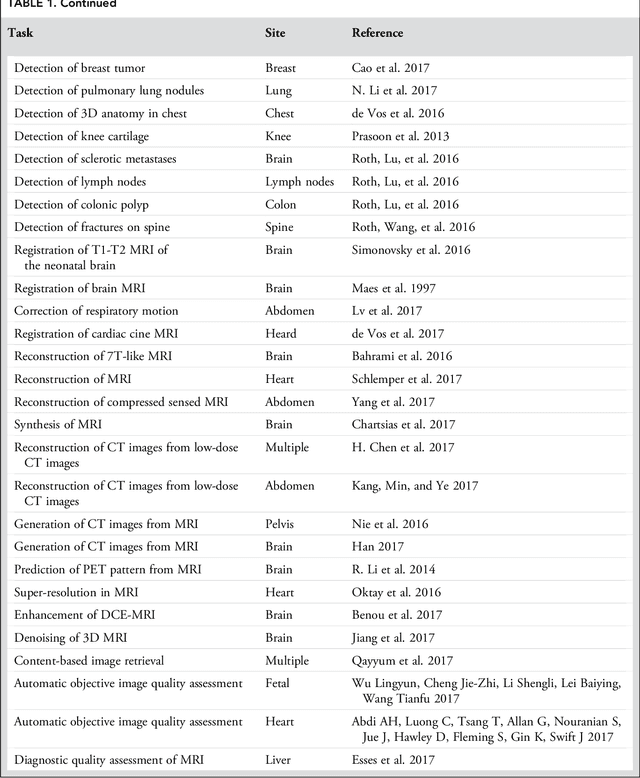
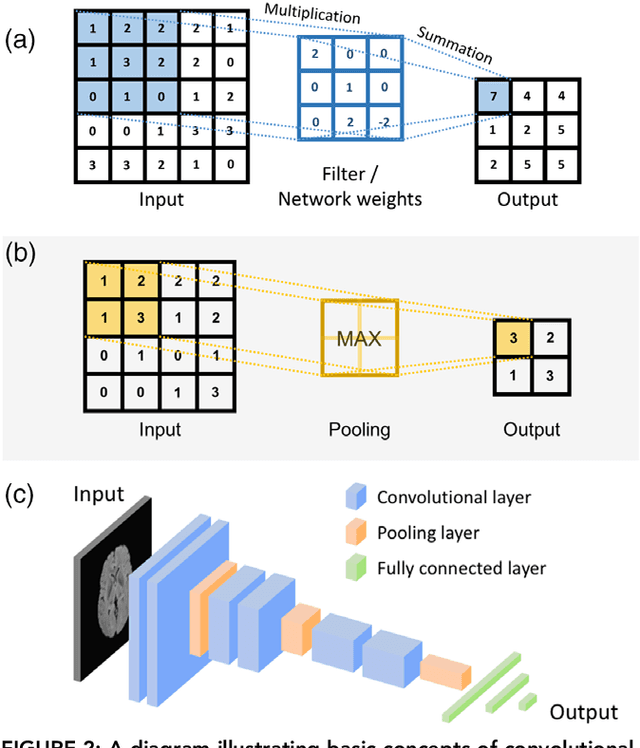
Abstract:Deep learning is a branch of artificial intelligence where networks of simple interconnected units are used to extract patterns from data in order to solve complex problems. Deep learning algorithms have shown groundbreaking performance in a variety of sophisticated tasks, especially those related to images. They have often matched or exceeded human performance. Since the medical field of radiology mostly relies on extracting useful information from images, it is a very natural application area for deep learning, and research in this area has rapidly grown in recent years. In this article, we review the clinical reality of radiology and discuss the opportunities for application of deep learning algorithms. We also introduce basic concepts of deep learning including convolutional neural networks. Then, we present a survey of the research in deep learning applied to radiology. We organize the studies by the types of specific tasks that they attempt to solve and review the broad range of utilized deep learning algorithms. Finally, we briefly discuss opportunities and challenges for incorporating deep learning in the radiology practice of the future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge