Mustafa R. Bashir
A personalized Uncertainty Quantification framework for patient survival models: estimating individual uncertainty of patients with metastatic brain tumors in the absence of ground truth
Nov 28, 2023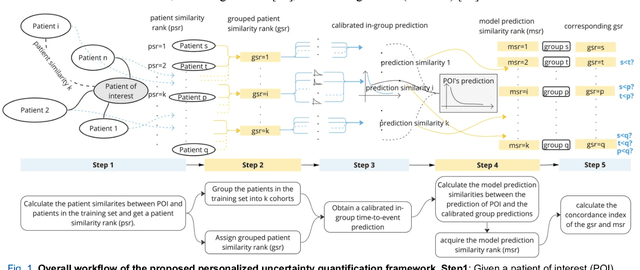
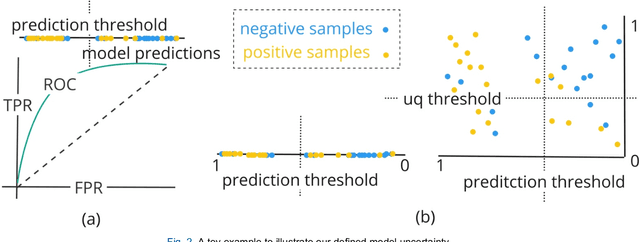
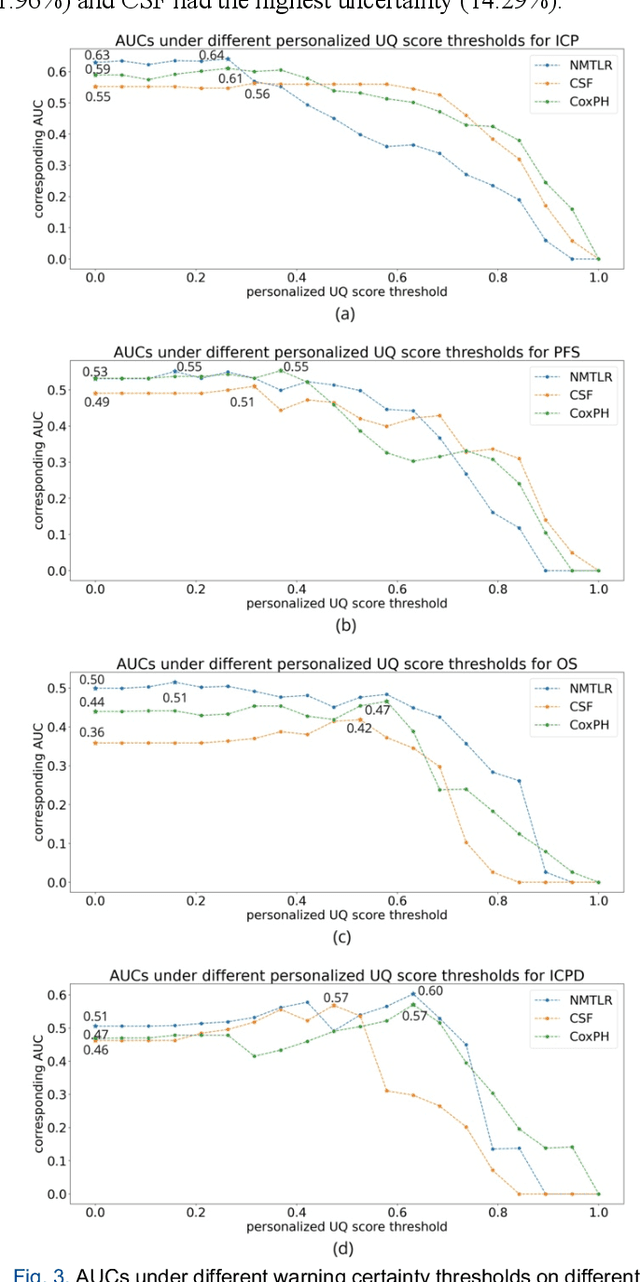
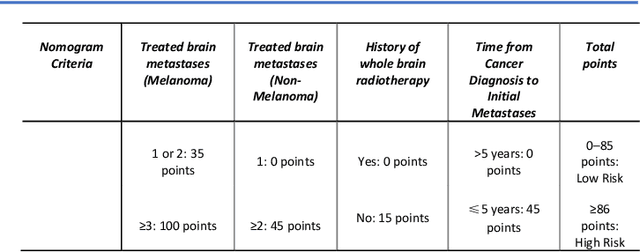
Abstract:TodevelopanovelUncertaintyQuantification (UQ) framework to estimate the uncertainty of patient survival models in the absence of ground truth, we developed and evaluated our approach based on a dataset of 1383 patients treated with stereotactic radiosurgery (SRS) for brain metastases between January 2015 and December 2020. Our motivating hypothesis is that a time-to-event prediction of a test patient on inference is more certain given a higher feature-space-similarity to patients in the training set. Therefore, the uncertainty for a particular patient-of-interest is represented by the concordance index between a patient similarity rank and a prediction similarity rank. Model uncertainty was defined as the increased percentage of the max uncertainty-constrained-AUC compared to the model AUC. We evaluated our method on multiple clinically-relevant endpoints, including time to intracranial progression (ICP), progression-free survival (PFS) after SRS, overall survival (OS), and time to ICP and/or death (ICPD), on a variety of both statistical and non-statistical models, including CoxPH, conditional survival forest (CSF), and neural multi-task linear regression (NMTLR). Our results show that all models had the lowest uncertainty on ICP (2.21%) and the highest uncertainty (17.28%) on ICPD. OS models demonstrated high variation in uncertainty performance, where NMTLR had the lowest uncertainty(1.96%)and CSF had the highest uncertainty (14.29%). In conclusion, our method can estimate the uncertainty of individual patient survival modeling results. As expected, our data empirically demonstrate that as model uncertainty measured via our technique increases, the similarity between a feature-space and its predicted outcome decreases.
Duke Spleen Data Set: A Publicly Available Spleen MRI and CT dataset for Training Segmentation
May 09, 2023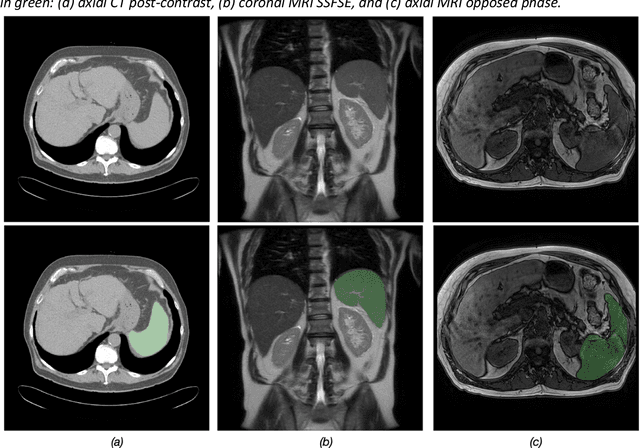

Abstract:Spleen volumetry is primarily associated with patients suffering from chronic liver disease and portal hypertension, as they often have spleens with abnormal shapes and sizes. However, manually segmenting the spleen to obtain its volume is a time-consuming process. Deep learning algorithms have proven to be effective in automating spleen segmentation, but a suitable dataset is necessary for training such algorithms. To our knowledge, the few publicly available datasets for spleen segmentation lack confounding features such as ascites and abdominal varices. To address this issue, the Duke Spleen Data Set (DSDS) has been developed, which includes 109 CT and MRI volumes from patients with chronic liver disease and portal hypertension. The dataset includes a diverse range of image types, vendors, planes, and contrasts, as well as varying spleen shapes and sizes due to underlying disease states. The DSDS aims to facilitate the creation of robust spleen segmentation models that can take into account these variations and confounding factors.
Deep learning in radiology: an overview of the concepts and a survey of the state of the art
Feb 10, 2018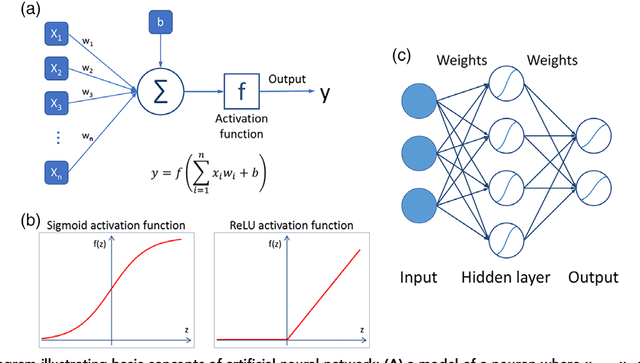
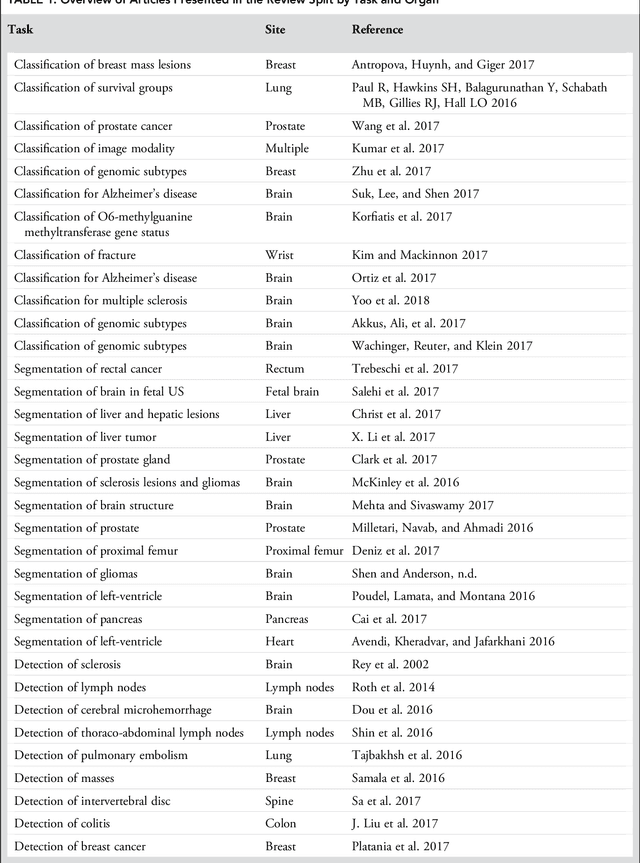
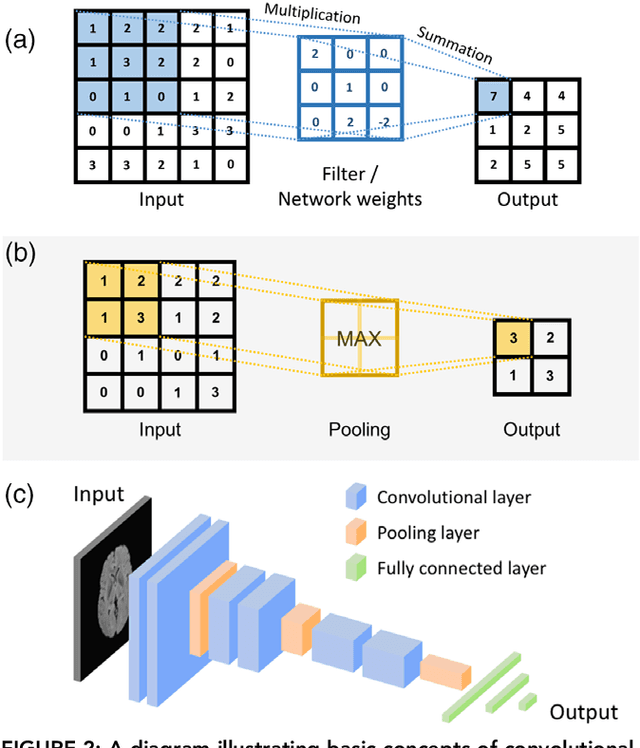
Abstract:Deep learning is a branch of artificial intelligence where networks of simple interconnected units are used to extract patterns from data in order to solve complex problems. Deep learning algorithms have shown groundbreaking performance in a variety of sophisticated tasks, especially those related to images. They have often matched or exceeded human performance. Since the medical field of radiology mostly relies on extracting useful information from images, it is a very natural application area for deep learning, and research in this area has rapidly grown in recent years. In this article, we review the clinical reality of radiology and discuss the opportunities for application of deep learning algorithms. We also introduce basic concepts of deep learning including convolutional neural networks. Then, we present a survey of the research in deep learning applied to radiology. We organize the studies by the types of specific tasks that they attempt to solve and review the broad range of utilized deep learning algorithms. Finally, we briefly discuss opportunities and challenges for incorporating deep learning in the radiology practice of the future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge