Benjamin Wildman-Tobriner
Diagnostic Impact of Cine Clips for Thyroid Nodule Assessment on Ultrasound
Feb 01, 2026Abstract:Background: Thyroid ultrasound is commonly performed using a combination of static images and cine clips (video recordings). However, the exact utility and impact of cine images remains unknown. This study aimed to evaluate the impact of cine imaging on accuracy and consistency of thyroid nodule assessment, using the American College of Radiology Thyroid Reporting and Data System (ACR TI-RADS). Methods: 50 benign and 50 malignant thyroid nodules with cytopathology results were included. A reader study with 4 specialty-trained radiologists was then conducted over 3 rounds, assessing only static images in the first two rounds and both static and cine images in the third round. TI-RADS scores and the consequent management recommendations were then evaluated by comparing them to the malignancy status of the nodules. Results: Mean sensitivity for malignancy detection was 0.65 for static images and 0.67 with both static and cine images (p>0.5). Specificity was 0.20 for static images and 0.22 with both static and cine images (p>0.5). Management recommendations were similar with and without cine images. Intrareader agreement on feature assignments remained consistent across all rounds, though TI-RADS point totals were slightly higher with cine images. Conclusion: The inclusion of cine imaging for thyroid nodule assessment on ultrasound did not significantly change diagnostic performance. Current practice guidelines, which do not mandate cine imaging, are sufficient for accurate diagnosis.
Effectiveness of Automatically Curated Dataset in Thyroid Nodules Classification Algorithms Using Deep Learning
Feb 01, 2026Abstract:The diagnosis of thyroid nodule cancers commonly utilizes ultrasound images. Several studies showed that deep learning algorithms designed to classify benign and malignant thyroid nodules could match radiologists' performance. However, data availability for training deep learning models is often limited due to the significant effort required to curate such datasets. The previous study proposed a method to curate thyroid nodule datasets automatically. It was tested to have a 63% yield rate and 83% accuracy. However, the usefulness of the generated data for training deep learning models remains unknown. In this study, we conducted experiments to determine whether using a automatically-curated dataset improves deep learning algorithms' performance. We trained deep learning models on the manually annotated and automatically-curated datasets. We also trained with a smaller subset of the automatically-curated dataset that has higher accuracy to explore the optimum usage of such dataset. As a result, the deep learning model trained on the manually selected dataset has an AUC of 0.643 (95% confidence interval [CI]: 0.62, 0.66). It is significantly lower than the AUC of the 6automatically-curated dataset trained deep learning model, 0.694 (95% confidence interval [CI]: 0.67, 0.73, P < .001). The AUC of the accurate subset trained deep learning model is 0.689 (95% confidence interval [CI]: 0.66, 0.72, P > .43), which is insignificantly worse than the AUC of the full automatically-curated dataset. In conclusion, we showed that using a automatically-curated dataset can substantially increase the performance of deep learning algorithms, and it is suggested to use all the data rather than only using the accurate subset.
Deep Learning for Classification of Thyroid Nodules on Ultrasound: Validation on an Independent Dataset
Jul 27, 2022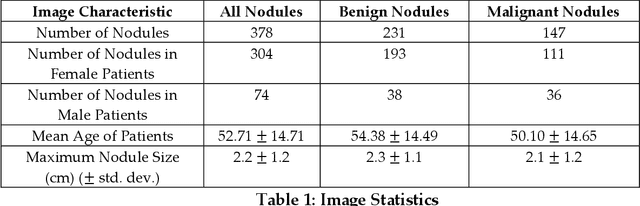
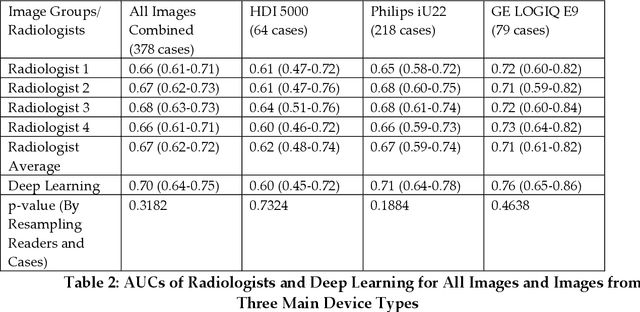
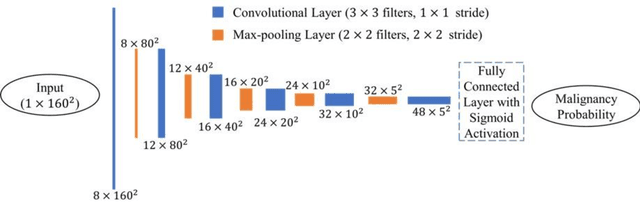
Abstract:Objectives: The purpose is to apply a previously validated deep learning algorithm to a new thyroid nodule ultrasound image dataset and compare its performances with radiologists. Methods: Prior study presented an algorithm which is able to detect thyroid nodules and then make malignancy classifications with two ultrasound images. A multi-task deep convolutional neural network was trained from 1278 nodules and originally tested with 99 separate nodules. The results were comparable with that of radiologists. The algorithm was further tested with 378 nodules imaged with ultrasound machines from different manufacturers and product types than the training cases. Four experienced radiologists were requested to evaluate the nodules for comparison with deep learning. Results: The Area Under Curve (AUC) of the deep learning algorithm and four radiologists were calculated with parametric, binormal estimation. For the deep learning algorithm, the AUC was 0.70 (95% CI: 0.64 - 0.75). The AUC of radiologists were 0.66 (95% CI: 0.61 - 0.71), 0.67 (95% CI:0.62 - 0.73), 0.68 (95% CI: 0.63 - 0.73), and 0.66 (95%CI: 0.61 - 0.71). Conclusion: In the new testing dataset, the deep learning algorithm achieved similar performances with all four radiologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge