Mark Michalski
DeepAAA: clinically applicable and generalizable detection of abdominal aortic aneurysm using deep learning
Jul 04, 2019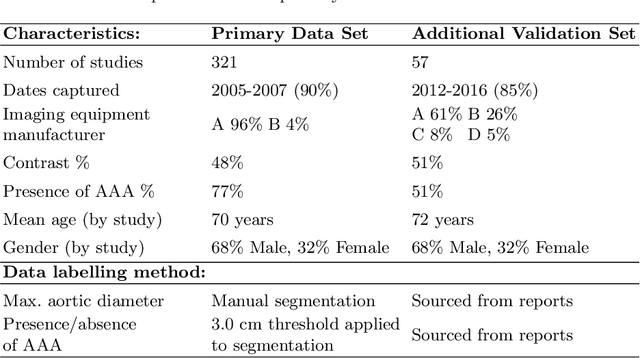
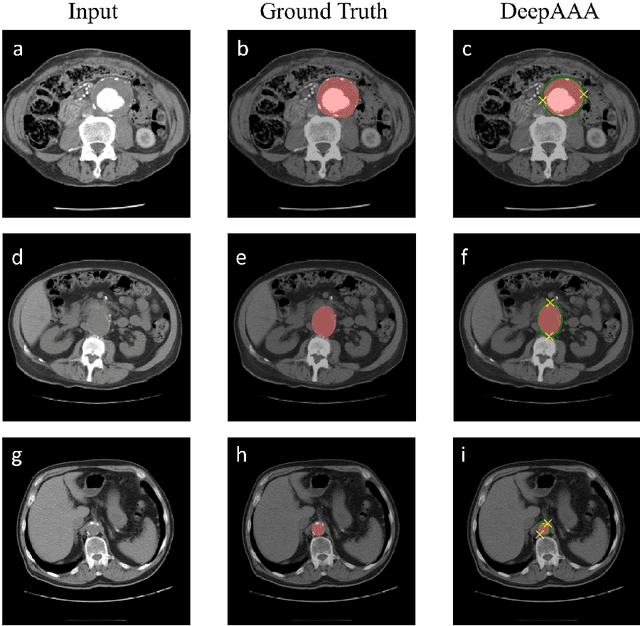
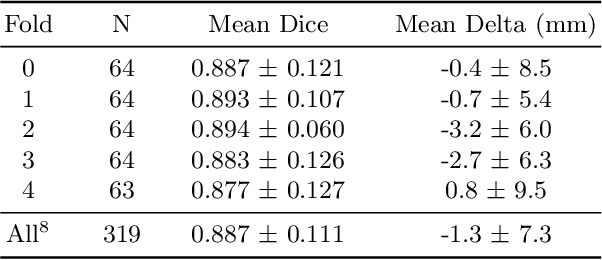
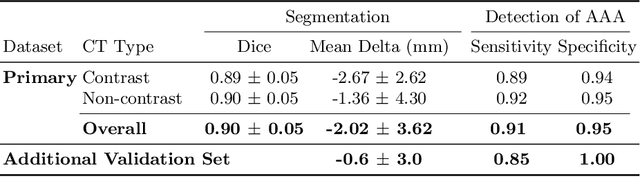
Abstract:We propose a deep learning-based technique for detection and quantification of abdominal aortic aneurysms (AAAs). The condition, which leads to more than 10,000 deaths per year in the United States, is asymptomatic, often detected incidentally, and often missed by radiologists. Our model architecture is a modified 3D U-Net combined with ellipse fitting that performs aorta segmentation and AAA detection. The study uses 321 abdominal-pelvic CT examinations performed by Massachusetts General Hospital Department of Radiology for training and validation. The model is then further tested for generalizability on a separate set of 57 examinations with differing patient demographics and acquisition characteristics than the original dataset. DeepAAA achieves high performance on both sets of data (sensitivity/specificity 0.91/0.95 and 0.85 / 1.0 respectively), on contrast and non-contrast CT scans and works with image volumes with varying numbers of images. We find that DeepAAA exceeds literature-reported performance of radiologists on incidental AAA detection. It is expected that the model can serve as an effective background detector in routine CT examinations to prevent incidental AAAs from being missed.
4D CNN for semantic segmentation of cardiac volumetric sequences
Jun 17, 2019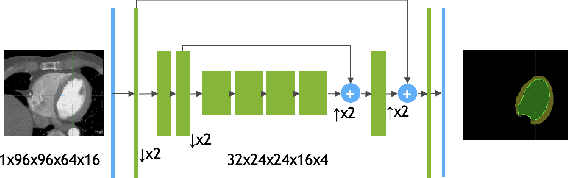
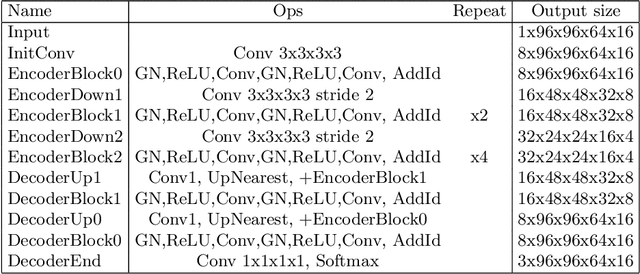
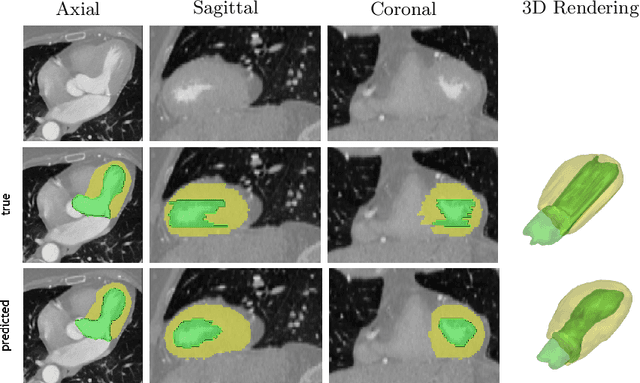
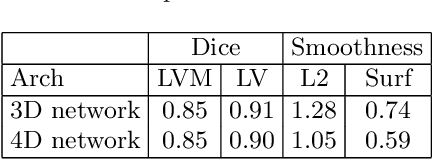
Abstract:We propose a 4D convolutional neural network (CNN) for the segmentation of retrospective ECG-gated cardiac CT, a series of single-channel volumetric data over time. While only a small subset of volumes in the temporal sequence are annotated, we define a sparse loss function on available labels to allow the network to leverage unlabeled images during training and generate a fully segmented sequence. We investigate the accuracy of the proposed 4D network to predict temporally consistent segmentations and compare with traditional 3D segmentation approaches. We demonstrate the feasibility of the 4D CNN and establish its performance on cardiac 4D CCTA.
Medical Image Synthesis for Data Augmentation and Anonymization using Generative Adversarial Networks
Sep 13, 2018
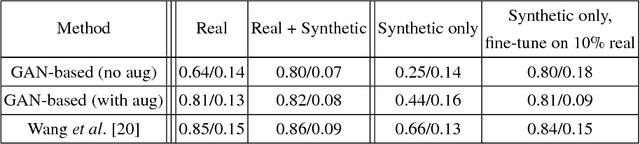
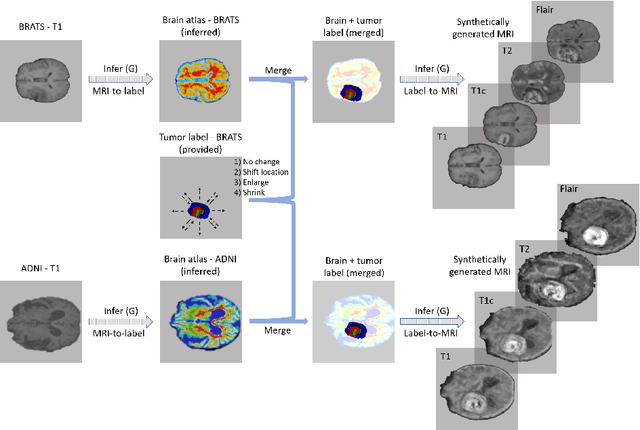

Abstract:Data diversity is critical to success when training deep learning models. Medical imaging data sets are often imbalanced as pathologic findings are generally rare, which introduces significant challenges when training deep learning models. In this work, we propose a method to generate synthetic abnormal MRI images with brain tumors by training a generative adversarial network using two publicly available data sets of brain MRI. We demonstrate two unique benefits that the synthetic images provide. First, we illustrate improved performance on tumor segmentation by leveraging the synthetic images as a form of data augmentation. Second, we demonstrate the value of generative models as an anonymization tool, achieving comparable tumor segmentation results when trained on the synthetic data versus when trained on real subject data. Together, these results offer a potential solution to two of the largest challenges facing machine learning in medical imaging, namely the small incidence of pathological findings, and the restrictions around sharing of patient data.
Fully-Automated Analysis of Body Composition from CT in Cancer Patients Using Convolutional Neural Networks
Aug 11, 2018



Abstract:The amounts of muscle and fat in a person's body, known as body composition, are correlated with cancer risks, cancer survival, and cardiovascular risk. The current gold standard for measuring body composition requires time-consuming manual segmentation of CT images by an expert reader. In this work, we describe a two-step process to fully automate the analysis of CT body composition using a DenseNet to select the CT slice and U-Net to perform segmentation. We train and test our methods on independent cohorts. Our results show Dice scores (0.95-0.98) and correlation coefficients (R=0.99) that are favorable compared to human readers. These results suggest that fully automated body composition analysis is feasible, which could enable both clinical use and large-scale population studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge