Liming Pan
Advancing Opinion Dynamics Modeling with Neural Diffusion-Convection-Reaction Equation
Feb 05, 2026Abstract:Advanced opinion dynamics modeling is vital for deciphering social behavior, emphasizing its role in mitigating polarization and securing cyberspace. To synergize mechanistic interpretability with data-driven flexibility, recent studies have explored the integration of Physics-Informed Neural Networks (PINNs) for opinion modeling. Despite this promise, existing methods are tailored to incomplete priors, lacking a comprehensive physical system to integrate dynamics from local, global, and endogenous levels. Moreover, penalty-based constraints adopted in existing methods struggle to deeply encode physical priors, leading to optimization pathologies and discrepancy between latent representations and physical transparency. To this end, we offer a physical view to interpret opinion dynamics via Diffusion-Convection-Reaction (DCR) system inspired by interacting particle theory. Building upon the Neural ODEs, we define the neural opinion dynamics to coordinate neural networks with physical priors, and further present the OPINN, a physics-informed neural framework for opinion dynamics modeling. Evaluated on real-world and synthetic datasets, OPINN achieves state-of-the-art performance in opinion evolution forecasting, offering a promising paradigm for the nexus of cyber, physical, and social systems.
TopoDIM: One-shot Topology Generation of Diverse Interaction Modes for Multi-Agent Systems
Jan 15, 2026Abstract:Optimizing communication topology in LLM-based multi-agent system is critical for enabling collective intelligence. Existing methods mainly rely on spatio-temporal interaction paradigms, where the sequential execution of multi-round dialogues incurs high latency and computation. Motivated by the recent insights that evaluation and debate mechanisms can improve problem-solving in multi-agent systems, we propose TopoDIM, a framework for one-shot Topology generation with Diverse Interaction Modes. Designed for decentralized execution to enhance adaptability and privacy, TopoDIM enables agents to autonomously construct heterogeneous communication without iterative coordination, achieving token efficiency and improved task performance. Experiments demonstrate that TopoDIM reduces total token consumption by 46.41% while improving average performance by 1.50% over state-of-the-art methods. Moreover, the framework exhibits strong adaptability in organizing communication among heterogeneous agents. Code is available at: https://anonymous.4open.science/r/TopoDIM-8D35/
EpiLLM: Unlocking the Potential of Large Language Models in Epidemic Forecasting
May 19, 2025Abstract:Advanced epidemic forecasting is critical for enabling precision containment strategies, highlighting its strategic importance for public health security. While recent advances in Large Language Models (LLMs) have demonstrated effectiveness as foundation models for domain-specific tasks, their potential for epidemic forecasting remains largely unexplored. In this paper, we introduce EpiLLM, a novel LLM-based framework tailored for spatio-temporal epidemic forecasting. Considering the key factors in real-world epidemic transmission: infection cases and human mobility, we introduce a dual-branch architecture to achieve fine-grained token-level alignment between such complex epidemic patterns and language tokens for LLM adaptation. To unleash the multi-step forecasting and generalization potential of LLM architectures, we propose an autoregressive modeling paradigm that reformulates the epidemic forecasting task into next-token prediction. To further enhance LLM perception of epidemics, we introduce spatio-temporal prompt learning techniques, which strengthen forecasting capabilities from a data-driven perspective. Extensive experiments show that EpiLLM significantly outperforms existing baselines on real-world COVID-19 datasets and exhibits scaling behavior characteristic of LLMs.
AutoCas: Autoregressive Cascade Predictor in Social Networks via Large Language Models
Feb 25, 2025Abstract:Popularity prediction in information cascades plays a crucial role in social computing, with broad applications in viral marketing, misinformation control, and content recommendation. However, information propagation mechanisms, user behavior, and temporal activity patterns exhibit significant diversity, necessitating a foundational model capable of adapting to such variations. At the same time, the amount of available cascade data remains relatively limited compared to the vast datasets used for training large language models (LLMs). Recent studies have demonstrated the feasibility of leveraging LLMs for time-series prediction by exploiting commonalities across different time-series domains. Building on this insight, we introduce the Autoregressive Information Cascade Predictor (AutoCas), an LLM-enhanced model designed specifically for cascade popularity prediction. Unlike natural language sequences, cascade data is characterized by complex local topologies, diffusion contexts, and evolving dynamics, requiring specialized adaptations for effective LLM integration. To address these challenges, we first tokenize cascade data to align it with sequence modeling principles. Next, we reformulate cascade diffusion as an autoregressive modeling task to fully harness the architectural strengths of LLMs. Beyond conventional approaches, we further introduce prompt learning to enhance the synergy between LLMs and cascade prediction. Extensive experiments demonstrate that AutoCas significantly outperforms baseline models in cascade popularity prediction while exhibiting scaling behavior inherited from LLMs. Code is available at this repository: https://anonymous.4open.science/r/AutoCas-85C6
A spring-block theory of feature learning in deep neural networks
Jul 28, 2024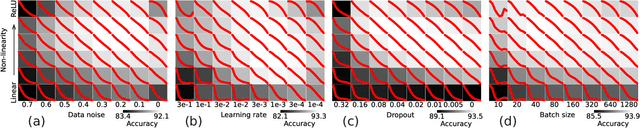
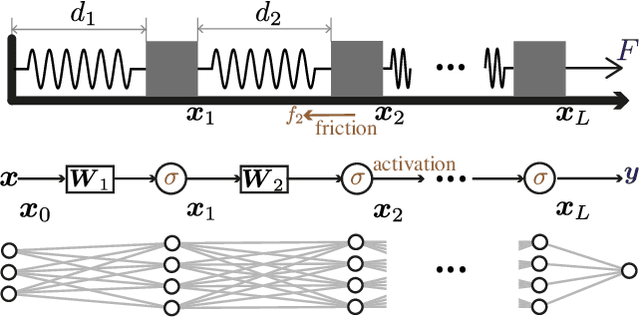
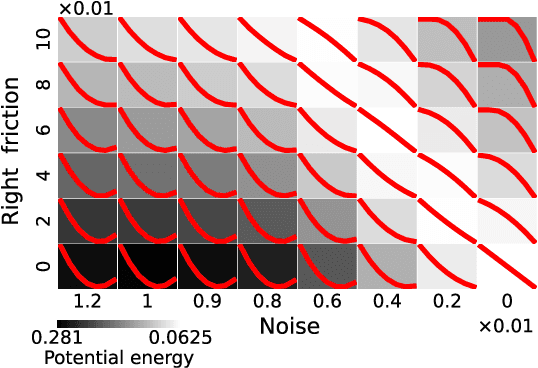
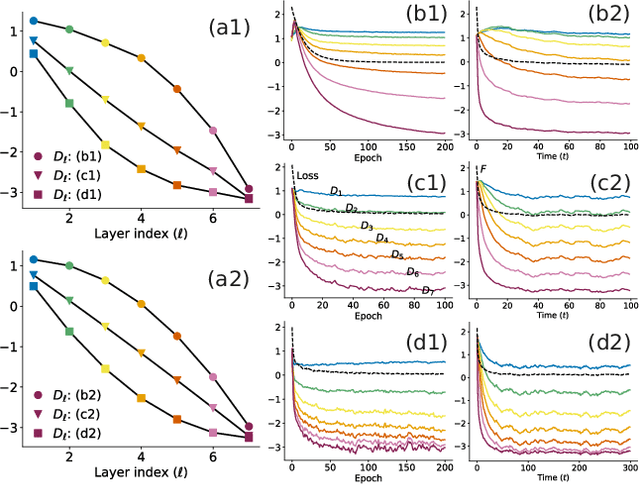
Abstract:A central question in deep learning is how deep neural networks (DNNs) learn features. DNN layers progressively collapse data into a regular low-dimensional geometry. This collective effect of non-linearity, noise, learning rate, width, depth, and numerous other parameters, has eluded first-principles theories which are built from microscopic neuronal dynamics. Here we present a noise-non-linearity phase diagram that highlights where shallow or deep layers learn features more effectively. We then propose a macroscopic mechanical theory of feature learning that accurately reproduces this phase diagram, offering a clear intuition for why and how some DNNs are ``lazy'' and some are ``active'', and relating the distribution of feature learning over layers with test accuracy.
A Dynamical Graph Prior for Relational Inference
Jun 09, 2023Abstract:Relational inference aims to identify interactions between parts of a dynamical system from the observed dynamics. Current state-of-the-art methods fit a graph neural network (GNN) on a learnable graph to the dynamics. They use one-step message-passing GNNs -- intuitively the right choice since non-locality of multi-step or spectral GNNs may confuse direct and indirect interactions. But the \textit{effective} interaction graph depends on the sampling rate and it is rarely localized to direct neighbors, leading to local minima for the one-step model. In this work, we propose a \textit{dynamical graph prior} (DYGR) for relational inference. The reason we call it a prior is that, contrary to established practice, it constructively uses error amplification in high-degree non-local polynomial filters to generate good gradients for graph learning. To deal with non-uniqueness, DYGR simultaneously fits a ``shallow'' one-step model with shared graph topology. Experiments show that DYGR reconstructs graphs far more accurately than earlier methods, with remarkable robustness to under-sampling. Since appropriate sampling rates for unknown dynamical systems are not known a priori, this robustness makes DYGR suitable for real applications in scientific machine learning.
Statistical Mechanics of Generalization In Graph Convolution Networks
Dec 26, 2022Abstract:Graph neural networks (GNN) have become the default machine learning model for relational datasets, including protein interaction networks, biological neural networks, and scientific collaboration graphs. We use tools from statistical physics and random matrix theory to precisely characterize generalization in simple graph convolution networks on the contextual stochastic block model. The derived curves are phenomenologically rich: they explain the distinction between learning on homophilic and heterophilic graphs and they predict double descent whose existence in GNNs has been questioned by recent work. Our results are the first to accurately explain the behavior not only of a stylized graph learning model but also of complex GNNs on messy real-world datasets. To wit, we use our analytic insights about homophily and heterophily to improve performance of state-of-the-art graph neural networks on several heterophilic benchmarks by a simple addition of negative self-loop filters.
Neural Link Prediction with Walk Pooling
Oct 08, 2021



Abstract:Graph neural networks achieve high accuracy in link prediction by jointly leveraging graph topology and node attributes. Topology, however, is represented indirectly; state-of-the-art methods based on subgraph classification label nodes with distance to the target link, so that, although topological information is present, it is tempered by pooling. This makes it challenging to leverage features like loops and motifs associated with network formation mechanisms. We propose a link prediction algorithm based on a new pooling scheme called WalkPool. WalkPool combines the expressivity of topological heuristics with the feature-learning ability of neural networks. It summarizes a putative link by random walk probabilities of adjacent paths. Instead of extracting transition probabilities from the original graph, it computes the transition matrix of a "predictive" latent graph by applying attention to learned features; this may be interpreted as feature-sensitive topology fingerprinting. WalkPool can leverage unsupervised node features or be combined with GNNs and trained end-to-end. It outperforms state-of-the-art methods on all common link prediction benchmarks, both homophilic and heterophilic, with and without node attributes. Applying WalkPool to a set of unsupervised GNNs significantly improves prediction accuracy, suggesting that it may be used as a general-purpose graph pooling scheme.
Link Prediction via Matrix Completion
Jun 23, 2016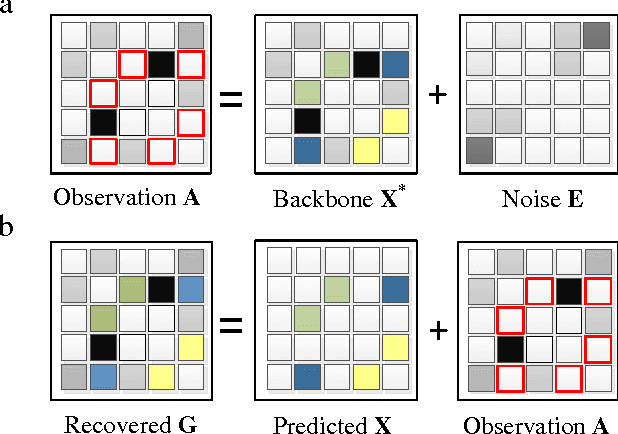
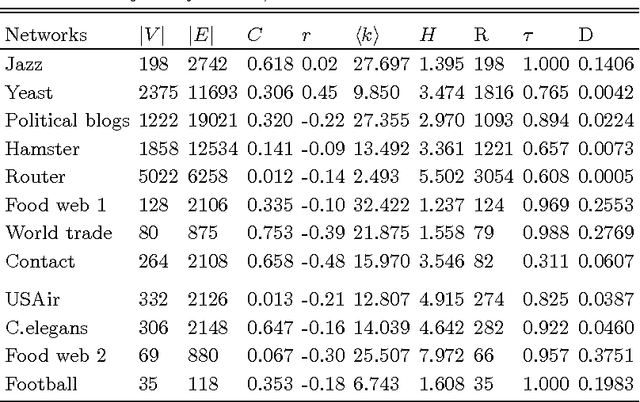
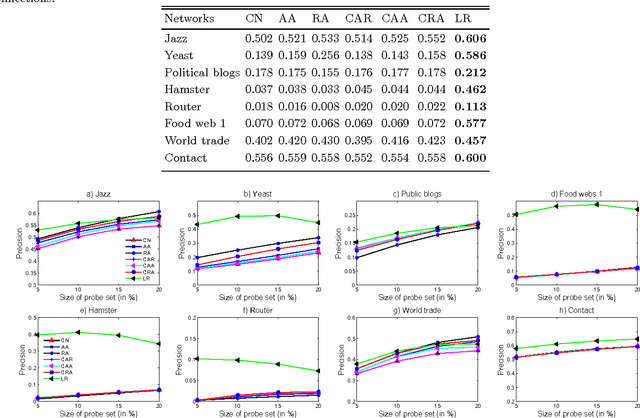
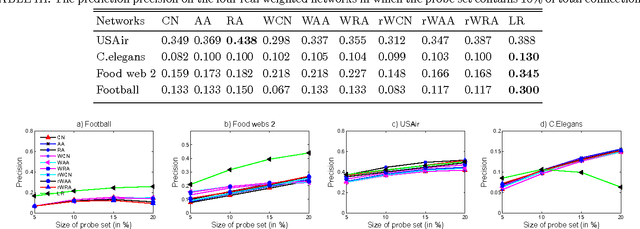
Abstract:Inspired by practical importance of social networks, economic networks, biological networks and so on, studies on large and complex networks have attracted a surge of attentions in the recent years. Link prediction is a fundamental issue to understand the mechanisms by which new links are added to the networks. We introduce the method of robust principal component analysis (robust PCA) into link prediction, and estimate the missing entries of the adjacency matrix. On one hand, our algorithm is based on the sparsity and low rank property of the matrix, on the other hand, it also performs very well when the network is dense. This is because a relatively dense real network is also sparse in comparison to the complete graph. According to extensive experiments on real networks from disparate fields, when the target network is connected and sufficiently dense, whatever it is weighted or unweighted, our method is demonstrated to be very effective and with prediction accuracy being considerably improved comparing with many state-of-the-art algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge