Kristoffer H. Madsen
Probabilistic Spatial Transformers for Bayesian Data Augmentation
Apr 07, 2020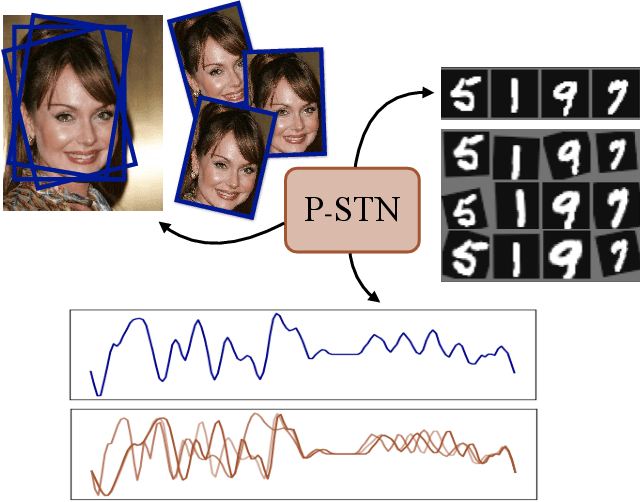
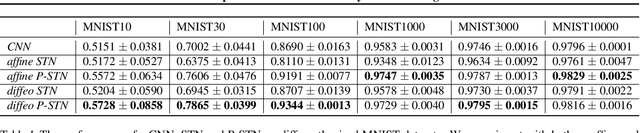
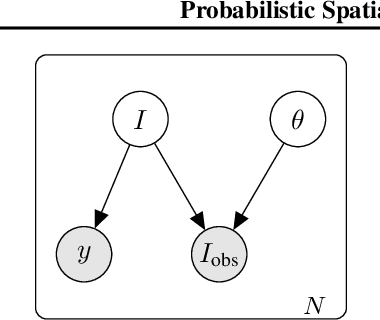
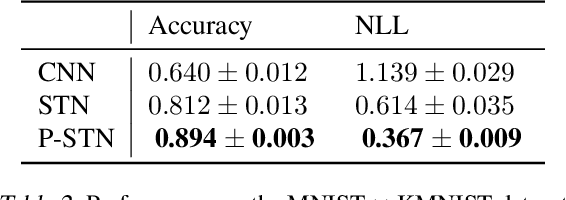
Abstract:High-capacity models require vast amounts of data, and data augmentation is a common remedy when this resource is limited. Standard augmentation techniques apply small hand-tuned transformations to existing data, which is a brittle process that realistically only allows for simple transformations. We propose a Bayesian interpretation of data augmentation where the transformations are modelled as latent variables to be marginalized, and show how these can be inferred variationally in an end-to-end fashion. This allows for significantly more complex transformations than manual tuning, and the marginalization implies a form of test-time data augmentation. The resulting model can be interpreted as a probabilistic extension of spatial transformer networks. Experimentally, we demonstrate improvements in accuracy and uncertainty quantification in image and time series classification tasks.
Probabilistic PARAFAC2
Jun 21, 2018



Abstract:The PARAFAC2 is a multimodal factor analysis model suitable for analyzing multi-way data when one of the modes has incomparable observation units, for example because of differences in signal sampling or batch sizes. A fully probabilistic treatment of the PARAFAC2 is desirable in order to improve robustness to noise and provide a well founded principle for determining the number of factors, but challenging because the factor loadings are constrained to be orthogonal. We develop two probabilistic formulations of the PARAFAC2 along with variational procedures for inference: In the one approach, the mean values of the factor loadings are orthogonal leading to closed form variational updates, and in the other, the factor loadings themselves are orthogonal using a matrix Von Mises-Fisher distribution. We contrast our probabilistic formulation to the conventional direct fitting algorithm based on maximum likelihood. On simulated data and real fluorescence spectroscopy and gas chromatography-mass spectrometry data, we compare our approach to the conventional PARAFAC2 model estimation and find that the probabilistic formulation is more robust to noise and model order misspecification. The probabilistic PARAFAC2 thus forms a promising framework for modeling multi-way data accounting for uncertainty.
Deep Convolutional Neural Networks for Interpretable Analysis of EEG Sleep Stage Scoring
Oct 02, 2017

Abstract:Sleep studies are important for diagnosing sleep disorders such as insomnia, narcolepsy or sleep apnea. They rely on manual scoring of sleep stages from raw polisomnography signals, which is a tedious visual task requiring the workload of highly trained professionals. Consequently, research efforts to purse for an automatic stage scoring based on machine learning techniques have been carried out over the last years. In this work, we resort to multitaper spectral analysis to create visually interpretable images of sleep patterns from EEG signals as inputs to a deep convolutional network trained to solve visual recognition tasks. As a working example of transfer learning, a system able to accurately classify sleep stages in new unseen patients is presented. Evaluations in a widely-used publicly available dataset favourably compare to state-of-the-art results, while providing a framework for visual interpretation of outcomes.
Scalable Group Level Probabilistic Sparse Factor Analysis
Dec 14, 2016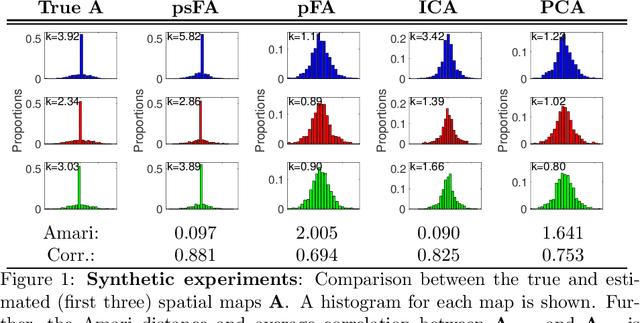

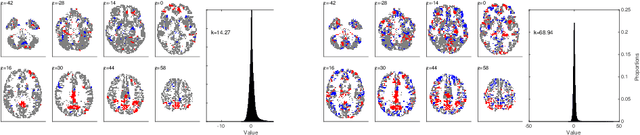
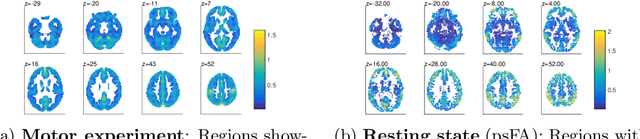
Abstract:Many data-driven approaches exist to extract neural representations of functional magnetic resonance imaging (fMRI) data, but most of them lack a proper probabilistic formulation. We propose a group level scalable probabilistic sparse factor analysis (psFA) allowing spatially sparse maps, component pruning using automatic relevance determination (ARD) and subject specific heteroscedastic spatial noise modeling. For task-based and resting state fMRI, we show that the sparsity constraint gives rise to components similar to those obtained by group independent component analysis. The noise modeling shows that noise is reduced in areas typically associated with activation by the experimental design. The psFA model identifies sparse components and the probabilistic setting provides a natural way to handle parameter uncertainties. The variational Bayesian framework easily extends to more complex noise models than the presently considered.
Nonparametric Modeling of Dynamic Functional Connectivity in fMRI Data
Jun 08, 2016
Abstract:Dynamic functional connectivity (FC) has in recent years become a topic of interest in the neuroimaging community. Several models and methods exist for both functional magnetic resonance imaging (fMRI) and electroencephalography (EEG), and the results point towards the conclusion that FC exhibits dynamic changes. The existing approaches modeling dynamic connectivity have primarily been based on time-windowing the data and k-means clustering. We propose a non-parametric generative model for dynamic FC in fMRI that does not rely on specifying window lengths and number of dynamic states. Rooted in Bayesian statistical modeling we use the predictive likelihood to investigate if the model can discriminate between a motor task and rest both within and across subjects. We further investigate what drives dynamic states using the model on the entire data collated across subjects and task/rest. We find that the number of states extracted are driven by subject variability and preprocessing differences while the individual states are almost purely defined by either task or rest. This questions how we in general interpret dynamic FC and points to the need for more research on what drives dynamic FC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge