Krishna Gadepalli
Human-centric Metric for Accelerating Pathology Reports Annotation
Nov 12, 2019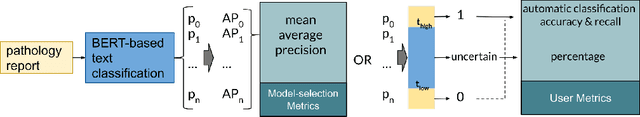
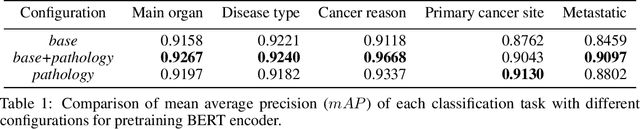
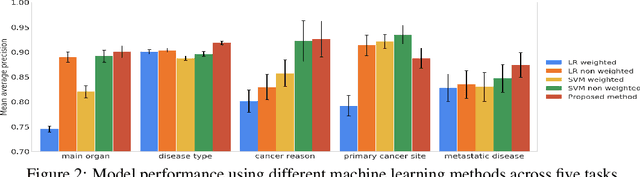
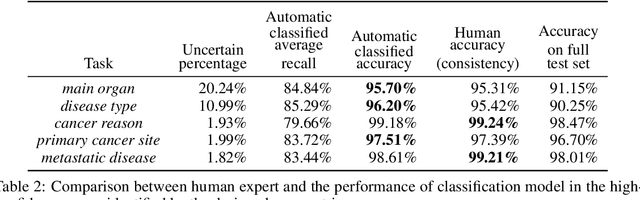
Abstract:Pathology reports contain useful information such as the main involved organ, diagnosis, etc. These information can be identified from the free text reports and used for large-scale statistical analysis or serve as annotation for other modalities such as pathology slides images. However, manual classification for a huge number of reports on multiple tasks is labor-intensive. In this paper, we have developed an automatic text classifier based on BERT and we propose a human-centric metric to evaluate the model. According to the model confidence, we identify low-confidence cases that require further expert annotation and high-confidence cases that are automatically classified. We report the percentage of low-confidence cases and the performance of automatically classified cases. On the high-confidence cases, the model achieves classification accuracy comparable to pathologists. This leads a potential of reducing 80% to 98% of the manual annotation workload.
Microscope 2.0: An Augmented Reality Microscope with Real-time Artificial Intelligence Integration
Dec 04, 2018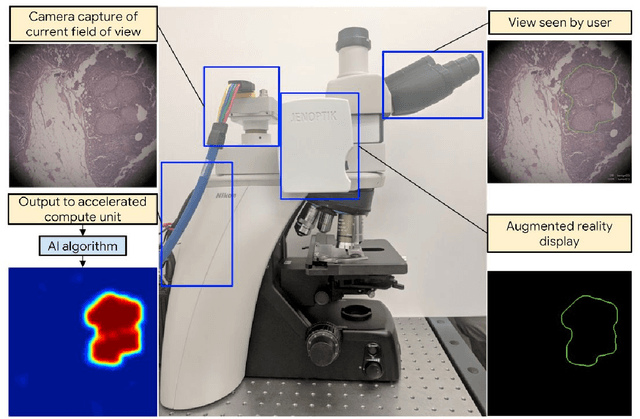
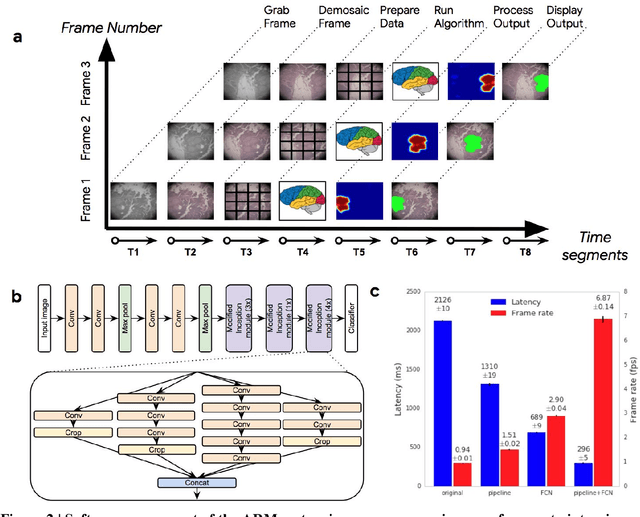
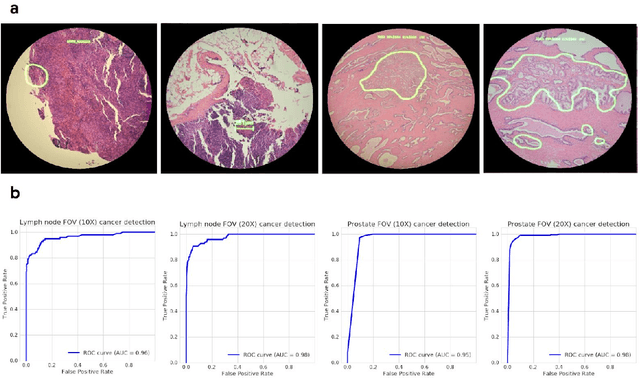
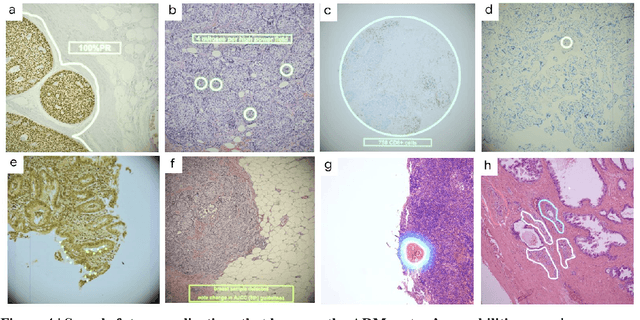
Abstract:The brightfield microscope is instrumental in the visual examination of both biological and physical samples at sub-millimeter scales. One key clinical application has been in cancer histopathology, where the microscopic assessment of the tissue samples is used for the diagnosis and staging of cancer and thus guides clinical therapy. However, the interpretation of these samples is inherently subjective, resulting in significant diagnostic variability. Moreover, in many regions of the world, access to pathologists is severely limited due to lack of trained personnel. In this regard, Artificial Intelligence (AI) based tools promise to improve the access and quality of healthcare. However, despite significant advances in AI research, integration of these tools into real-world cancer diagnosis workflows remains challenging because of the costs of image digitization and difficulties in deploying AI solutions. Here we propose a cost-effective solution to the integration of AI: the Augmented Reality Microscope (ARM). The ARM overlays AI-based information onto the current view of the sample through the optical pathway in real-time, enabling seamless integration of AI into the regular microscopy workflow. We demonstrate the utility of ARM in the detection of lymph node metastases in breast cancer and the identification of prostate cancer with a latency that supports real-time workflows. We anticipate that ARM will remove barriers towards the use of AI in microscopic analysis and thus improve the accuracy and efficiency of cancer diagnosis. This approach is applicable to other microscopy tasks and AI algorithms in the life sciences and beyond.
Detecting Cancer Metastases on Gigapixel Pathology Images
Mar 08, 2017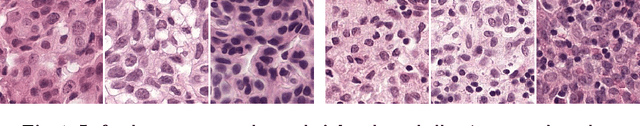

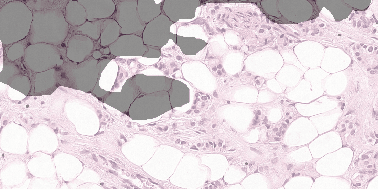
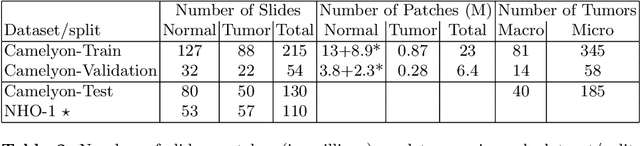
Abstract:Each year, the treatment decisions for more than 230,000 breast cancer patients in the U.S. hinge on whether the cancer has metastasized away from the breast. Metastasis detection is currently performed by pathologists reviewing large expanses of biological tissues. This process is labor intensive and error-prone. We present a framework to automatically detect and localize tumors as small as 100 x 100 pixels in gigapixel microscopy images sized 100,000 x 100,000 pixels. Our method leverages a convolutional neural network (CNN) architecture and obtains state-of-the-art results on the Camelyon16 dataset in the challenging lesion-level tumor detection task. At 8 false positives per image, we detect 92.4% of the tumors, relative to 82.7% by the previous best automated approach. For comparison, a human pathologist attempting exhaustive search achieved 73.2% sensitivity. We achieve image-level AUC scores above 97% on both the Camelyon16 test set and an independent set of 110 slides. In addition, we discover that two slides in the Camelyon16 training set were erroneously labeled normal. Our approach could considerably reduce false negative rates in metastasis detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge