Angela Lin
Human-centric Metric for Accelerating Pathology Reports Annotation
Nov 12, 2019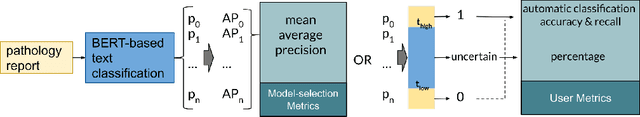
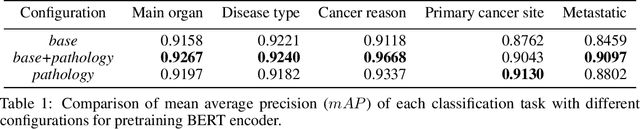
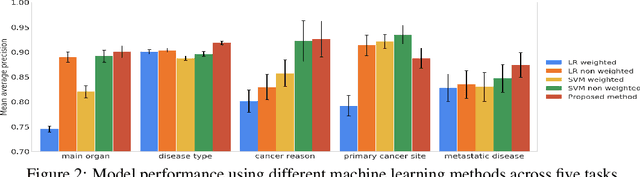
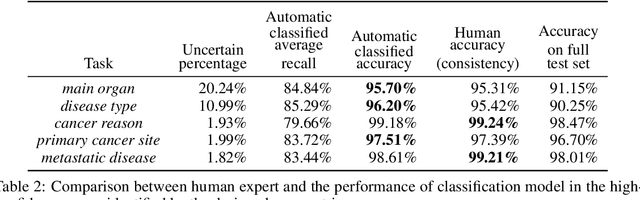
Abstract:Pathology reports contain useful information such as the main involved organ, diagnosis, etc. These information can be identified from the free text reports and used for large-scale statistical analysis or serve as annotation for other modalities such as pathology slides images. However, manual classification for a huge number of reports on multiple tasks is labor-intensive. In this paper, we have developed an automatic text classifier based on BERT and we propose a human-centric metric to evaluate the model. According to the model confidence, we identify low-confidence cases that require further expert annotation and high-confidence cases that are automatically classified. We report the percentage of low-confidence cases and the performance of automatically classified cases. On the high-confidence cases, the model achieves classification accuracy comparable to pathologists. This leads a potential of reducing 80% to 98% of the manual annotation workload.
Multimodal Multitask Representation Learning for Pathology Biobank Metadata Prediction
Sep 17, 2019



Abstract:Metadata are general characteristics of the data in a well-curated and condensed format, and have been proven to be useful for decision making, knowledge discovery, and also heterogeneous data organization of biobank. Among all data types in the biobank, pathology is the key component of the biobank and also serves as the gold standard of diagnosis. To maximize the utility of biobank and allow the rapid progress of biomedical science, it is essential to organize the data with well-populated pathology metadata. However, manual annotation of such information is tedious and time-consuming. In the study, we develop a multimodal multitask learning framework to predict four major slide-level metadata of pathology images. The framework learns generalizable representations across tissue slides, pathology reports, and case-level structured data. We demonstrate improved performance across all four tasks with the proposed method compared to a single modal single task baseline on two test sets, one external test set from a distinct data source (TCGA) and one internal held-out test set (TTH). In the test sets, the performance improvements on the averaged area under receiver operating characteristic curve across the four tasks are 16.48% and 9.05% on TCGA and TTH, respectively. Such pathology metadata prediction system may be adopted to mitigate the effort of expert annotation and ultimately accelerate the data-driven research by better utilization of the pathology biobank.
Improved Twitter Sentiment Analysis Using Naive Bayes and Custom Language Model
Nov 10, 2017



Abstract:In the last couple decades, social network services like Twitter have generated large volumes of data about users and their interests, providing meaningful business intelligence so organizations can better understand and engage their customers. All businesses want to know who is promoting their products, who is complaining about them, and how are these opinions bringing or diminishing value to a company. Companies want to be able to identify their high-value customers and quantify the value each user brings. Many businesses use social media metrics to calculate the user contribution score, which enables them to quantify the value that influential users bring on social media, so the businesses can offer them more differentiated services. However, the score calculation can be refined to provide a better illustration of a user's contribution. Using Microsoft Azure as a case study, we conducted Twitter sentiment analysis to develop a machine learning classification model that identifies tweet contents and sentiments most illustrative of positive-value user contribution. Using data mining and AI-powered cognitive tools, we analyzed factors of social influence and specifically, promotional language in the developer community. Our predictive model was a combination of a traditional supervised machine learning algorithm and a custom-developed natural language model for identifying promotional tweets, that identifies a product-specific promotion on Twitter with a 90% accuracy rate.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge