Joanna Wardlaw
Development of a Deep Learning Method to Identify Acute Ischemic Stroke Lesions on Brain CT
Sep 29, 2023



Abstract:Computed Tomography (CT) is commonly used to image acute ischemic stroke (AIS) patients, but its interpretation by radiologists is time-consuming and subject to inter-observer variability. Deep learning (DL) techniques can provide automated CT brain scan assessment, but usually require annotated images. Aiming to develop a DL method for AIS using labelled but not annotated CT brain scans from patients with AIS, we designed a convolutional neural network-based DL algorithm using routinely-collected CT brain scans from the Third International Stroke Trial (IST-3), which were not acquired using strict research protocols. The DL model aimed to detect AIS lesions and classify the side of the brain affected. We explored the impact of AIS lesion features, background brain appearances, and timing on DL performance. From 5772 unique CT scans of 2347 AIS patients (median age 82), 54% had visible AIS lesions according to expert labelling. Our best-performing DL method achieved 72% accuracy for lesion presence and side. Lesions that were larger (80% accuracy) or multiple (87% accuracy for two lesions, 100% for three or more), were better detected. Follow-up scans had 76% accuracy, while baseline scans 67% accuracy. Chronic brain conditions reduced accuracy, particularly non-stroke lesions and old stroke lesions (32% and 31% error rates respectively). DL methods can be designed for AIS lesion detection on CT using the vast quantities of routinely-collected CT brain scan data. Ultimately, this should lead to more robust and widely-applicable methods.
Challenges of building medical image datasets for development of deep learning software in stroke
Sep 26, 2023Abstract:Despite the large amount of brain CT data generated in clinical practice, the availability of CT datasets for deep learning (DL) research is currently limited. Furthermore, the data can be insufficiently or improperly prepared for machine learning and thus lead to spurious and irreproducible analyses. This lack of access to comprehensive and diverse datasets poses a significant challenge for the development of DL algorithms. In this work, we propose a complete semi-automatic pipeline to address the challenges of preparing a clinical brain CT dataset for DL analysis and describe the process of standardising this heterogeneous dataset. Challenges include handling image sets with different orientations (axial, sagittal, coronal), different image types (to view soft tissues or bones) and dimensions, and removing redundant background. The final pipeline was able to process 5,868/10,659 (45%) CT image datasets. Reasons for rejection include non-axial data (n=1,920), bone reformats (n=687), separated skull base/vault images (n=1,226), and registration failures (n=465). Further format adjustments, including image cropping, resizing and scaling are also needed for DL processing. Of the axial scans that were not localisers, bone reformats or split brains, 5,868/6,333 (93%) were accepted, while the remaining 465 failed the registration process. Appropriate preparation of medical imaging datasets for DL is a costly and time-intensive process.
Diffusion Models for Counterfactual Generation and Anomaly Detection in Brain Images
Aug 03, 2023



Abstract:Segmentation masks of pathological areas are useful in many medical applications, such as brain tumour and stroke management. Moreover, healthy counterfactuals of diseased images can be used to enhance radiologists' training files and to improve the interpretability of segmentation models. In this work, we present a weakly supervised method to generate a healthy version of a diseased image and then use it to obtain a pixel-wise anomaly map. To do so, we start by considering a saliency map that approximately covers the pathological areas, obtained with ACAT. Then, we propose a technique that allows to perform targeted modifications to these regions, while preserving the rest of the image. In particular, we employ a diffusion model trained on healthy samples and combine Denoising Diffusion Probabilistic Model (DDPM) and Denoising Diffusion Implicit Model (DDIM) at each step of the sampling process. DDPM is used to modify the areas affected by a lesion within the saliency map, while DDIM guarantees reconstruction of the normal anatomy outside of it. The two parts are also fused at each timestep, to guarantee the generation of a sample with a coherent appearance and a seamless transition between edited and unedited parts. We verify that when our method is applied to healthy samples, the input images are reconstructed without significant modifications. We compare our approach with alternative weakly supervised methods on IST-3 for stroke lesion segmentation and on BraTS2021 for brain tumour segmentation, where we improve the DICE score of the best competing method from $0.6534$ to $0.7056$.
ACAT: Adversarial Counterfactual Attention for Classification and Detection in Medical Imaging
Mar 27, 2023Abstract:In some medical imaging tasks and other settings where only small parts of the image are informative for the classification task, traditional CNNs can sometimes struggle to generalise. Manually annotated Regions of Interest (ROI) are sometimes used to isolate the most informative parts of the image. However, these are expensive to collect and may vary significantly across annotators. To overcome these issues, we propose a framework that employs saliency maps to obtain soft spatial attention masks that modulate the image features at different scales. We refer to our method as Adversarial Counterfactual Attention (ACAT). ACAT increases the baseline classification accuracy of lesions in brain CT scans from 71.39% to 72.55% and of COVID-19 related findings in lung CT scans from 67.71% to 70.84% and exceeds the performance of competing methods. We investigate the best way to generate the saliency maps employed in our architecture and propose a way to obtain them from adversarially generated counterfactual images. They are able to isolate the area of interest in brain and lung CT scans without using any manual annotations. In the task of localising the lesion location out of 6 possible regions, they obtain a score of 65.05% on brain CT scans, improving the score of 61.29% obtained with the best competing method.
GAN Augmentation: Augmenting Training Data using Generative Adversarial Networks
Oct 25, 2018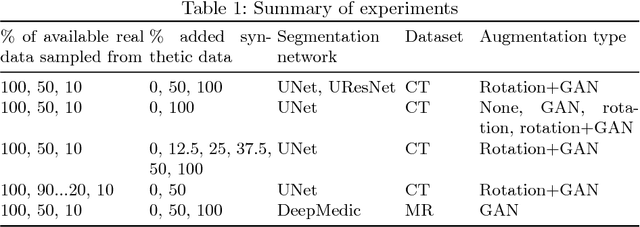
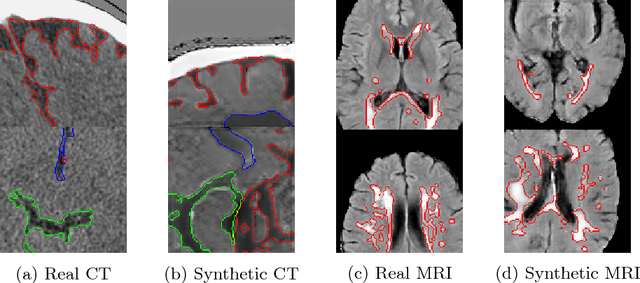
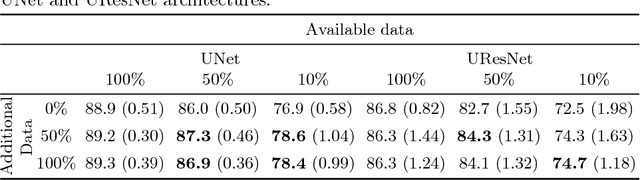
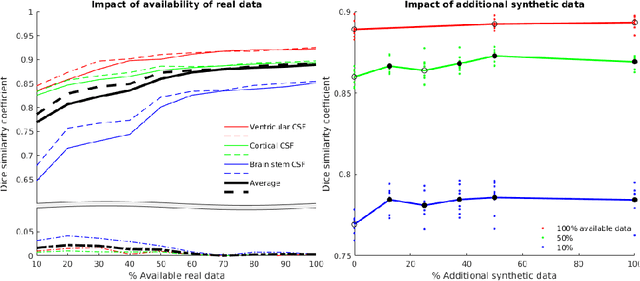
Abstract:One of the biggest issues facing the use of machine learning in medical imaging is the lack of availability of large, labelled datasets. The annotation of medical images is not only expensive and time consuming but also highly dependent on the availability of expert observers. The limited amount of training data can inhibit the performance of supervised machine learning algorithms which often need very large quantities of data on which to train to avoid overfitting. So far, much effort has been directed at extracting as much information as possible from what data is available. Generative Adversarial Networks (GANs) offer a novel way to unlock additional information from a dataset by generating synthetic samples with the appearance of real images. This paper demonstrates the feasibility of introducing GAN derived synthetic data to the training datasets in two brain segmentation tasks, leading to improvements in Dice Similarity Coefficient (DSC) of between 1 and 5 percentage points under different conditions, with the strongest effects seen fewer than ten training image stacks are available.
Machine learning of neuroimaging to diagnose cognitive impairment and dementia: a systematic review and comparative analysis
Apr 11, 2018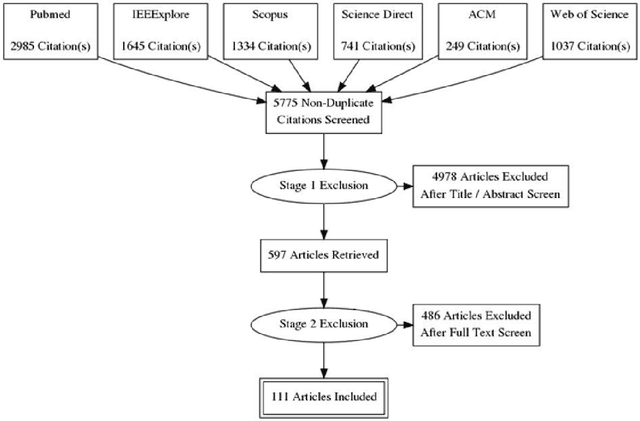
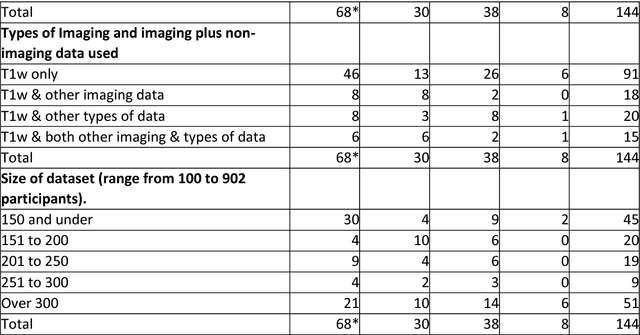
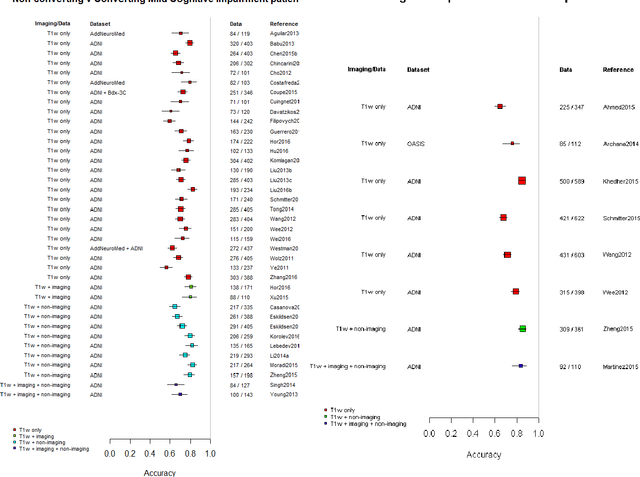
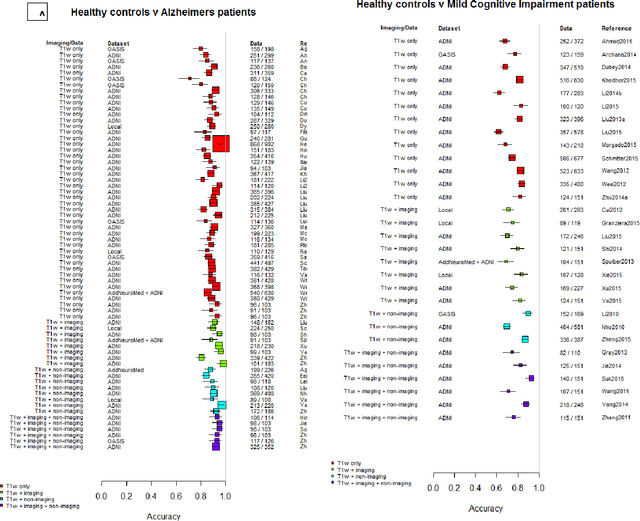
Abstract:INTRODUCTION: Advanced machine learning methods might help to identify dementia risk from neuroimaging, but their accuracy to date is unclear. METHODS: We systematically reviewed the literature, 2006 to late 2016, for machine learning studies differentiating healthy ageing through to dementia of various types, assessing study quality, and comparing accuracy at different disease boundaries. RESULTS: Of 111 relevant studies, most assessed Alzheimer's disease (AD) vs healthy controls, used ADNI data, support vector machines and only T1-weighted sequences. Accuracy was highest for differentiating AD from healthy controls, and poor for differentiating healthy controls vs MCI vs AD, or MCI converters vs non-converters. Accuracy increased using combined data types, but not by data source, sample size or machine learning method. DISCUSSION: Machine learning does not differentiate clinically-relevant disease categories yet. More diverse datasets, combinations of different types of data, and close clinical integration of machine learning would help to advance the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge