Jingsong Lv
GQWformer: A Quantum-based Transformer for Graph Representation Learning
Dec 03, 2024



Abstract:Graph Transformers (GTs) have demonstrated significant advantages in graph representation learning through their global attention mechanisms. However, the self-attention mechanism in GTs tends to neglect the inductive biases inherent in graph structures, making it chanllenging to effectively capture essential structural information. To address this issue, we propose a novel approach that integrate graph inductive bias into self-attention mechanisms by leveraging quantum technology for structural encoding. In this paper, we introduce the Graph Quantum Walk Transformer (GQWformer), a groundbreaking GNN framework that utilizes quantum walks on attributed graphs to generate node quantum states. These quantum states encapsulate rich structural attributes and serve as inductive biases for the transformer, thereby enabling the generation of more meaningful attention scores. By subsequently incorporating a recurrent neural network, our design amplifies the model's ability to focus on both local and global information. We conducted comprehensive experiments across five publicly available datasets to evaluate the effectiveness of our model. These results clearly indicate that GQWformer outperforms existing state-of-the-art graph classification algorithms. These findings highlight the significant potential of integrating quantum computing methodologies with traditional GNNs to advance the field of graph representation learning, providing a promising direction for future research and applications.
Improving Graph Out-of-distribution Generalization on Real-world Data
Jul 14, 2024



Abstract:Existing methods for graph out-of-distribution (OOD) generalization primarily rely on empirical studies on synthetic datasets. Such approaches tend to overemphasize the causal relationships between invariant sub-graphs and labels, thereby neglecting the non-negligible role of environment in real-world scenarios. In contrast to previous studies that impose rigid independence assumptions on environments and invariant sub-graphs, this paper presents the theorems of environment-label dependency and mutable rationale invariance, where the former characterizes the usefulness of environments in determining graph labels while the latter refers to the mutable importance of graph rationales. Based on analytic investigations, a novel variational inference based method named ``Probability Dependency on Environments and Rationales for OOD Graphs on Real-world Data'' (DEROG) is introduced. To alleviate the adverse effect of unknown prior knowledge on environments and rationales, DEROG utilizes generalized Bayesian inference. Further, DEROG employs an EM-based algorithm for optimization. Finally, extensive experiments on real-world datasets under different distribution shifts are conducted to show the superiority of DEROG. Our code is publicly available at https://anonymous.4open.science/r/DEROG-536B.
Learning Invariant Molecular Representation in Latent Discrete Space
Oct 22, 2023Abstract:Molecular representation learning lays the foundation for drug discovery. However, existing methods suffer from poor out-of-distribution (OOD) generalization, particularly when data for training and testing originate from different environments. To address this issue, we propose a new framework for learning molecular representations that exhibit invariance and robustness against distribution shifts. Specifically, we propose a strategy called ``first-encoding-then-separation'' to identify invariant molecule features in the latent space, which deviates from conventional practices. Prior to the separation step, we introduce a residual vector quantization module that mitigates the over-fitting to training data distributions while preserving the expressivity of encoders. Furthermore, we design a task-agnostic self-supervised learning objective to encourage precise invariance identification, which enables our method widely applicable to a variety of tasks, such as regression and multi-label classification. Extensive experiments on 18 real-world molecular datasets demonstrate that our model achieves stronger generalization against state-of-the-art baselines in the presence of various distribution shifts. Our code is available at https://github.com/HICAI-ZJU/iMoLD.
Circle Feature Graphormer: Can Circle Features Stimulate Graph Transformer?
Sep 11, 2023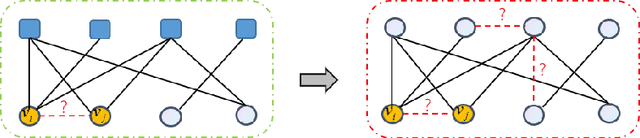
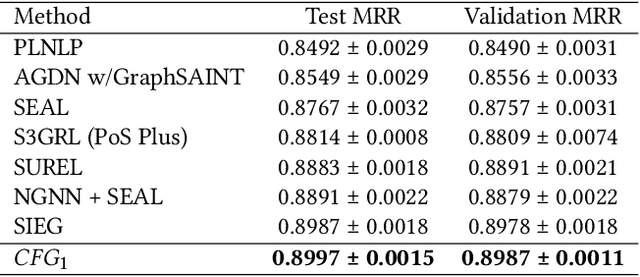
Abstract:In this paper, we introduce two local graph features for missing link prediction tasks on ogbl-citation2. We define the features as Circle Features, which are borrowed from the concept of circle of friends. We propose the detailed computing formulas for the above features. Firstly, we define the first circle feature as modified swing for common graph, which comes from bipartite graph. Secondly, we define the second circle feature as bridge, which indicates the importance of two nodes for different circle of friends. In addition, we firstly propose the above features as bias to enhance graph transformer neural network, such that graph self-attention mechanism can be improved. We implement a Circled Feature aware Graph transformer (CFG) model based on SIEG network, which utilizes a double tower structure to capture both global and local structure features. Experimental results show that CFG achieves the state-of-the-art performance on dataset ogbl-citation2.
Path-aware Siamese Graph Neural Network for Link Prediction
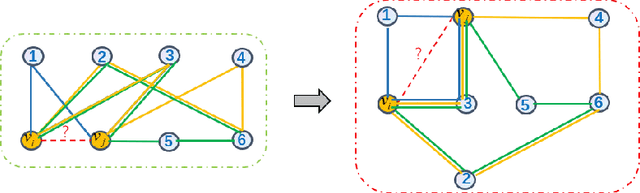
Aug 10, 2022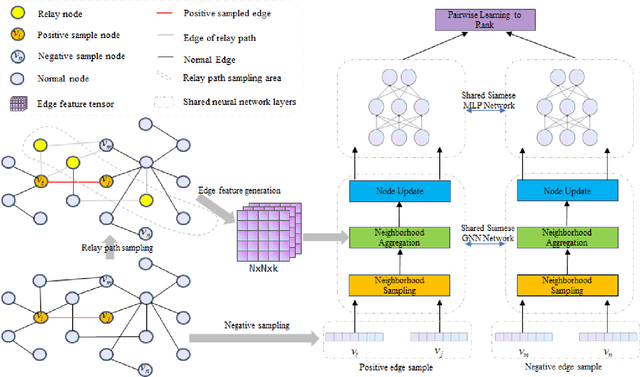
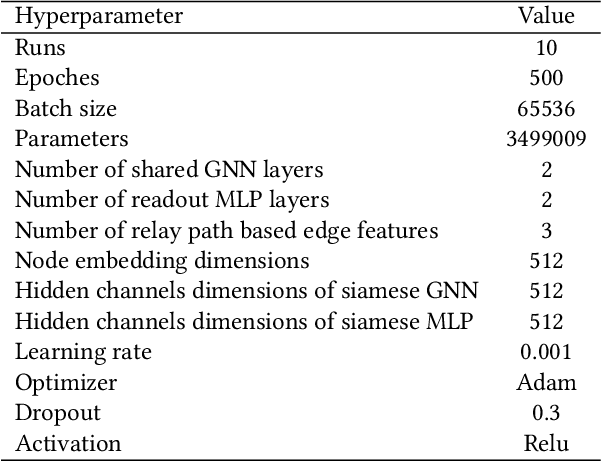
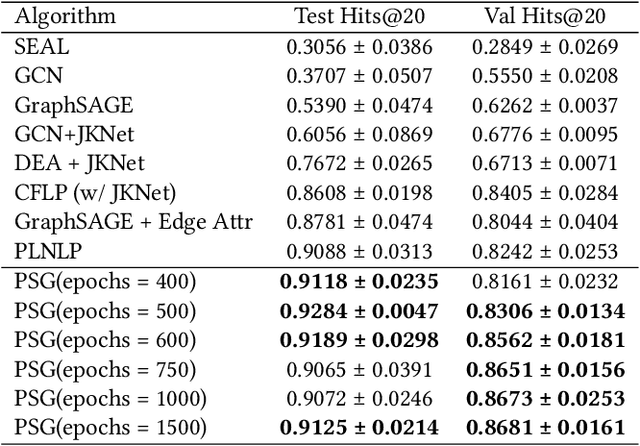
Abstract:In this paper, we propose an algorithm of Path-aware Siamese Graph neural network(PSG) for link prediction tasks. Firstly, PSG can capture both nodes and edge features for given two nodes, namely the structure information of k-neighborhoods and relay paths information of the nodes. Furthermore, siamese graph neural network is utilized by PSG for representation learning of two contrastive links, which are a positive link and a negative link. We evaluate the proposed algorithm PSG on a link property prediction dataset of Open Graph Benchmark (OGB), ogbl-ddi. PSG achieves top 1 performance on ogbl-ddi. The experimental results verify the superiority of PSG.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge