Haosen Wang
Adversarial Yet Cooperative: Multi-Perspective Reasoning in Retrieved-Augmented Language Models
Jan 08, 2026Abstract:Recent advances in synergizing large reasoning models (LRMs) with retrieval-augmented generation (RAG) have shown promising results, yet two critical challenges remain: (1) reasoning models typically operate from a single, unchallenged perspective, limiting their ability to conduct deep, self-correcting reasoning over external documents, and (2) existing training paradigms rely excessively on outcome-oriented rewards, which provide insufficient signal for shaping the complex, multi-step reasoning process. To address these issues, we propose an Reasoner-Verifier framework named Adversarial Reasoning RAG (ARR). The Reasoner and Verifier engage in reasoning on retrieved evidence and critiquing each other's logic while being guided by process-aware advantage that requires no external scoring model. This reward combines explicit observational signals with internal model uncertainty to jointly optimize reasoning fidelity and verification rigor. Experiments on multiple benchmarks demonstrate the effectiveness of our method.
Homophily-aware Heterogeneous Graph Contrastive Learning
Jan 15, 2025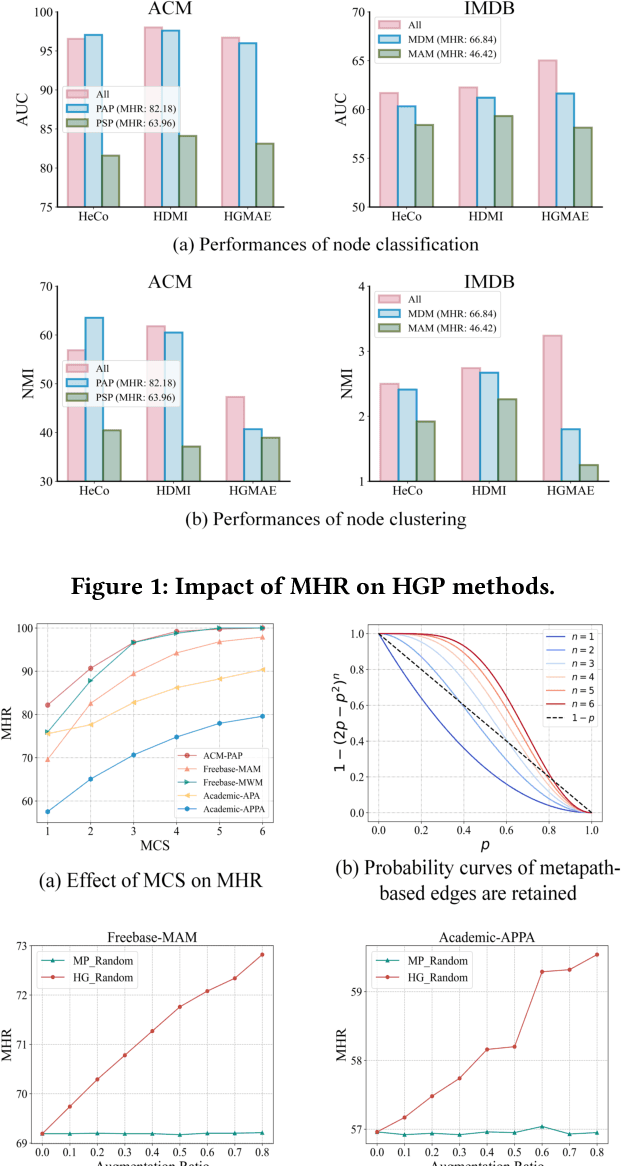
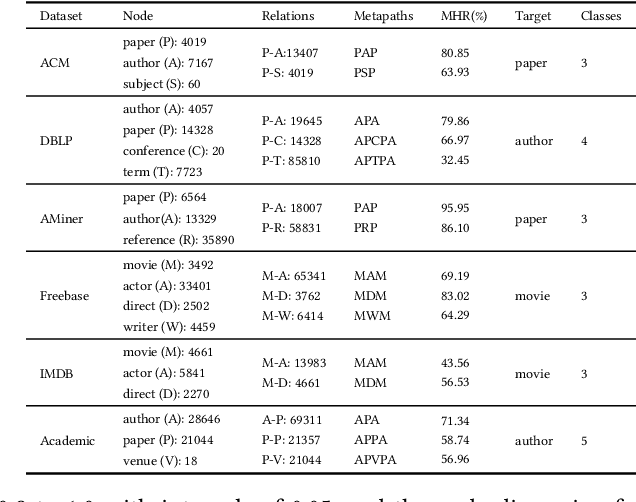
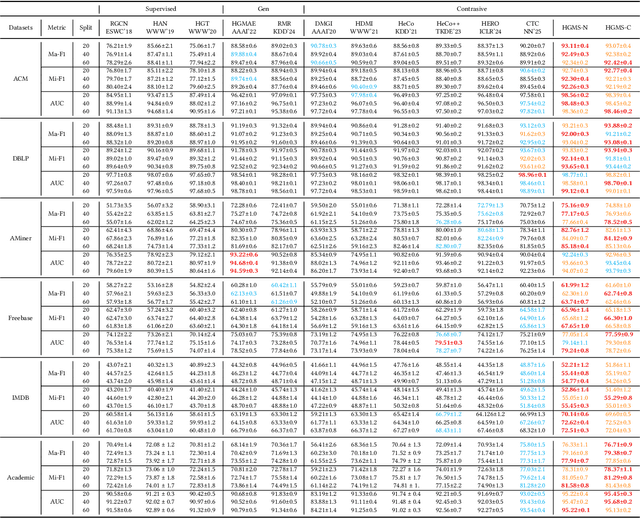
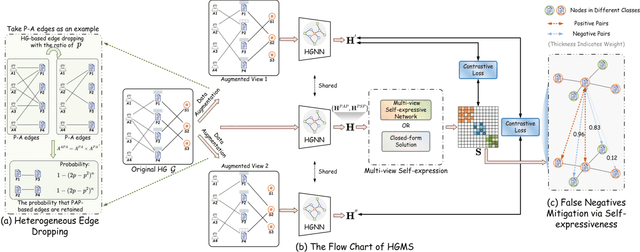
Abstract:Heterogeneous graph pre-training (HGP) has demonstrated remarkable performance across various domains. However, the issue of heterophily in real-world heterogeneous graphs (HGs) has been largely overlooked. To bridge this research gap, we proposed a novel heterogeneous graph contrastive learning framework, termed HGMS, which leverages connection strength and multi-view self-expression to learn homophilous node representations. Specifically, we design a heterogeneous edge dropping augmentation strategy that enhances the homophily of augmented views. Moreover, we introduce a multi-view self-expressive learning method to infer the homophily between nodes. In practice, we develop two approaches to solve the self-expressive matrix. The solved self-expressive matrix serves as an additional augmented view to provide homophilous information and is used to identify false negatives in contrastive loss. Extensive experimental results demonstrate the superiority of HGMS across different downstream tasks.
Improving Graph Out-of-distribution Generalization on Real-world Data
Jul 14, 2024



Abstract:Existing methods for graph out-of-distribution (OOD) generalization primarily rely on empirical studies on synthetic datasets. Such approaches tend to overemphasize the causal relationships between invariant sub-graphs and labels, thereby neglecting the non-negligible role of environment in real-world scenarios. In contrast to previous studies that impose rigid independence assumptions on environments and invariant sub-graphs, this paper presents the theorems of environment-label dependency and mutable rationale invariance, where the former characterizes the usefulness of environments in determining graph labels while the latter refers to the mutable importance of graph rationales. Based on analytic investigations, a novel variational inference based method named ``Probability Dependency on Environments and Rationales for OOD Graphs on Real-world Data'' (DEROG) is introduced. To alleviate the adverse effect of unknown prior knowledge on environments and rationales, DEROG utilizes generalized Bayesian inference. Further, DEROG employs an EM-based algorithm for optimization. Finally, extensive experiments on real-world datasets under different distribution shifts are conducted to show the superiority of DEROG. Our code is publicly available at https://anonymous.4open.science/r/DEROG-536B.
Geometric-Facilitated Denoising Diffusion Model for 3D Molecule Generation
Jan 05, 2024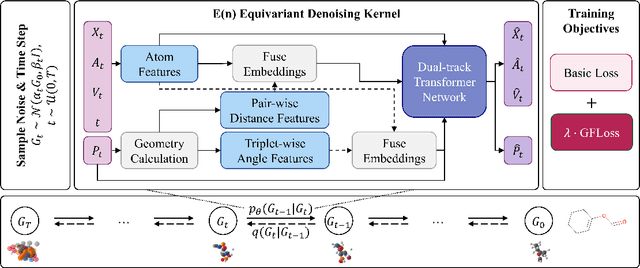
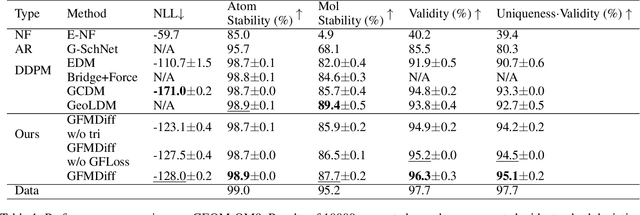
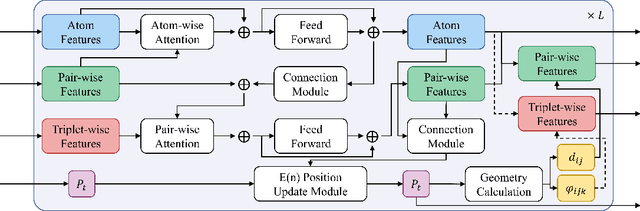
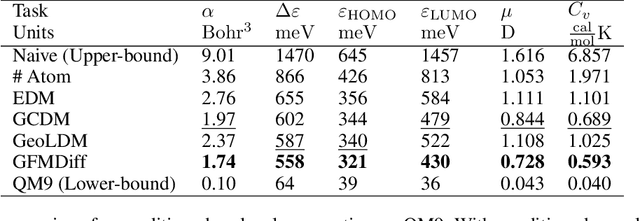
Abstract:Denoising diffusion models have shown great potential in multiple research areas. Existing diffusion-based generative methods on de novo 3D molecule generation face two major challenges. Since majority heavy atoms in molecules allow connections to multiple atoms through single bonds, solely using pair-wise distance to model molecule geometries is insufficient. Therefore, the first one involves proposing an effective neural network as the denoising kernel that is capable to capture complex multi-body interatomic relationships and learn high-quality features. Due to the discrete nature of graphs, mainstream diffusion-based methods for molecules heavily rely on predefined rules and generate edges in an indirect manner. The second challenge involves accommodating molecule generation to diffusion and accurately predicting the existence of bonds. In our research, we view the iterative way of updating molecule conformations in diffusion process is consistent with molecular dynamics and introduce a novel molecule generation method named Geometric-Facilitated Molecular Diffusion (GFMDiff). For the first challenge, we introduce a Dual-Track Transformer Network (DTN) to fully excevate global spatial relationships and learn high quality representations which contribute to accurate predictions of features and geometries. As for the second challenge, we design Geometric-Facilitated Loss (GFLoss) which intervenes the formation of bonds during the training period, instead of directly embedding edges into the latent space. Comprehensive experiments on current benchmarks demonstrate the superiority of GFMDiff.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge