James Taylor
Improving Audio Classification by Transitioning from Zero- to Few-Shot
Jul 26, 2025Abstract:State-of-the-art audio classification often employs a zero-shot approach, which involves comparing audio embeddings with embeddings from text describing the respective audio class. These embeddings are usually generated by neural networks trained through contrastive learning to align audio and text representations. Identifying the optimal text description for an audio class is challenging, particularly when the class comprises a wide variety of sounds. This paper examines few-shot methods designed to improve classification accuracy beyond the zero-shot approach. Specifically, audio embeddings are grouped by class and processed to replace the inherently noisy text embeddings. Our results demonstrate that few-shot classification typically outperforms the zero-shot baseline.
Event-based Motion-Robust Accurate Shape Estimation for Mixed Reflectance Scenes
Nov 16, 2023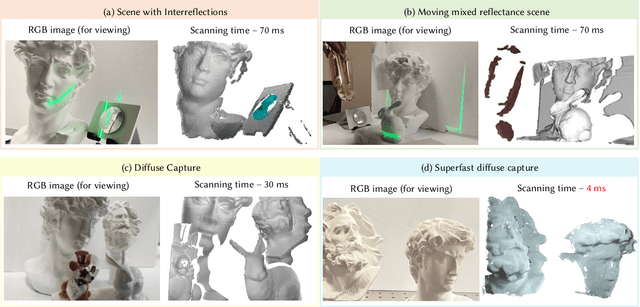
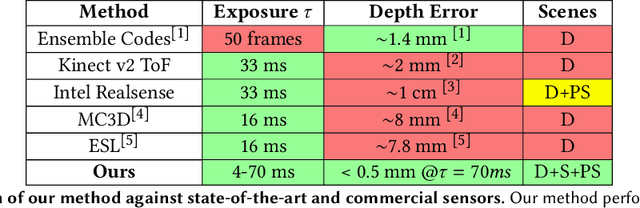
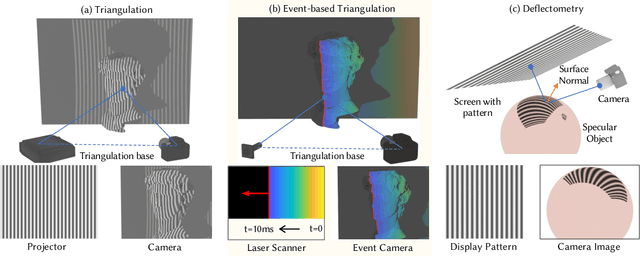
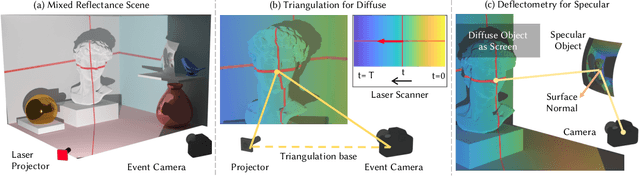
Abstract:Event-based structured light systems have recently been introduced as an exciting alternative to conventional frame-based triangulation systems for the 3D measurements of diffuse surfaces. Important benefits include the fast capture speed and the high dynamic range provided by the event camera - albeit at the cost of lower data quality. So far, both low-accuracy event-based as well as high-accuracy frame-based 3D imaging systems are tailored to a specific surface type, such as diffuse or specular, and can not be used for a broader class of object surfaces ("mixed reflectance scenes"). In this paper, we present a novel event-based structured light system that enables fast 3D imaging of mixed reflectance scenes with high accuracy. On the captured events, we use epipolar constraints that intrinsically enable decomposing the measured reflections into diffuse, two-bounce specular, and other multi-bounce reflections. The diffuse objects in the scene are reconstructed using triangulation. Eventually, the reconstructed diffuse scene parts are used as a "display" to evaluate the specular scene parts via deflectometry. This novel procedure allows us to use the entire scene as a virtual screen, using only a scanning laser and an event camera. The resulting system achieves fast and motion-robust (14Hz) reconstructions of mixed reflectance scenes with < 500 $\mu$m accuracy. Moreover, we introduce a "superfast" capture mode (250Hz) for the 3D measurement of diffuse scenes.
RxRx1: A Dataset for Evaluating Experimental Batch Correction Methods
Jan 13, 2023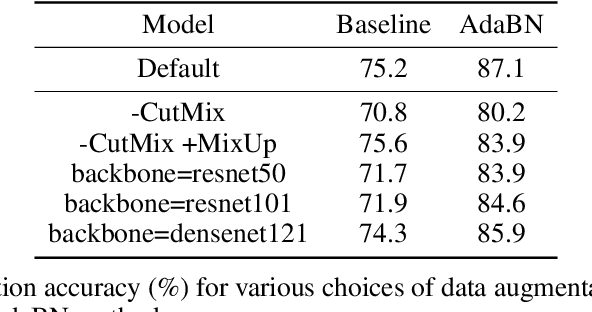



Abstract:High-throughput screening techniques are commonly used to obtain large quantities of data in many fields of biology. It is well known that artifacts arising from variability in the technical execution of different experimental batches within such screens confound these observations and can lead to invalid biological conclusions. It is therefore necessary to account for these batch effects when analyzing outcomes. In this paper we describe RxRx1, a biological dataset designed specifically for the systematic study of batch effect correction methods. The dataset consists of 125,510 high-resolution fluorescence microscopy images of human cells under 1,138 genetic perturbations in 51 experimental batches across 4 cell types. Visual inspection of the images alone clearly demonstrates significant batch effects. We propose a classification task designed to evaluate the effectiveness of experimental batch correction methods on these images and examine the performance of a number of correction methods on this task. Our goal in releasing RxRx1 is to encourage the development of effective experimental batch correction methods that generalize well to unseen experimental batches. The dataset can be downloaded at https://rxrx.ai.
Recurrent Kalman Networks: Factorized Inference in High-Dimensional Deep Feature Spaces
May 17, 2019

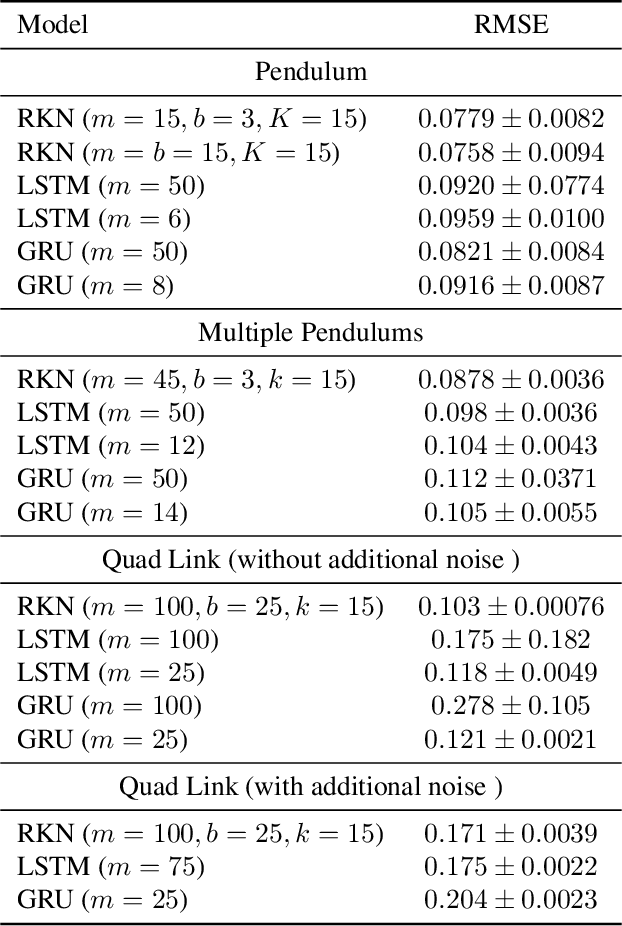
Abstract:In order to integrate uncertainty estimates into deep time-series modelling, Kalman Filters (KFs) (Kalman et al., 1960) have been integrated with deep learning models, however, such approaches typically rely on approximate inference techniques such as variational inference which makes learning more complex and often less scalable due to approximation errors. We propose a new deep approach to Kalman filtering which can be learned directly in an end-to-end manner using backpropagation without additional approximations. Our approach uses a high-dimensional factorized latent state representation for which the Kalman updates simplify to scalar operations and thus avoids hard to backpropagate, computationally heavy and potentially unstable matrix inversions. Moreover, we use locally linear dynamic models to efficiently propagate the latent state to the next time step. The resulting network architecture, which we call Recurrent Kalman Network (RKN), can be used for any time-series data, similar to a LSTM (Hochreiter & Schmidhuber, 1997) but uses an explicit representation of uncertainty. As shown by our experiments, the RKN obtains much more accurate uncertainty estimates than an LSTM or Gated Recurrent Units (GRUs) (Cho et al., 2014) while also showing a slightly improved prediction performance and outperforms various recent generative models on an image imputation task.
Hardware and Software manual for Evolution of Oil Droplets in a Chemo-Robotic Platform
Nov 07, 2014



Abstract:This manual outlines a fully automated liquid handling robot to enable physically-embodied evolution within a chemical oil-droplet system. The robot is based upon the REPRAP3D printer system and makes the droplets by mixing chemicals and then placing them in a petri dish after which they are recorded using a camera and the behaviour of the droplets analysed using image recognition software. This manual accompanies the open access publication published in Nature Communications DOI: 10.1038/ncomms6571.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge