Ishani Karmarkar
Subsampling Graphs with GNN Performance Guarantees
Feb 23, 2025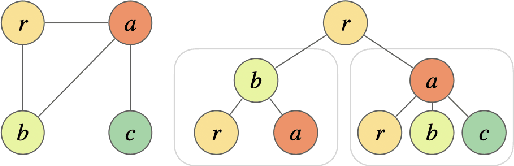
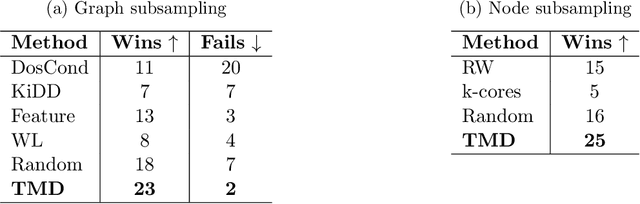
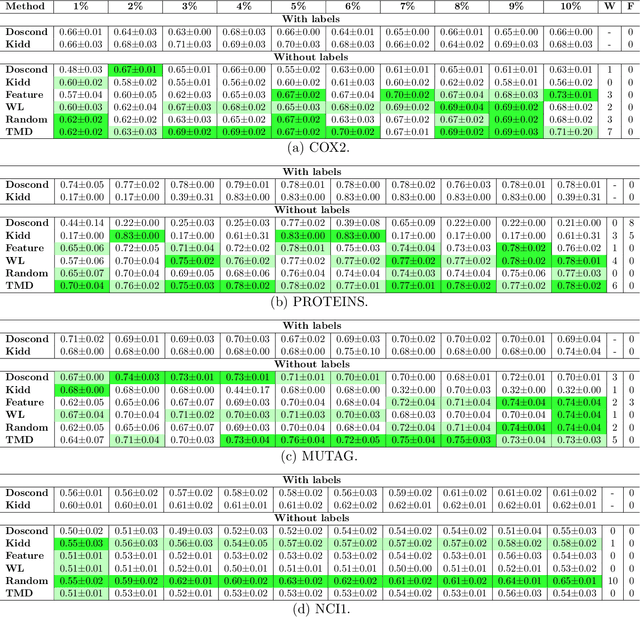
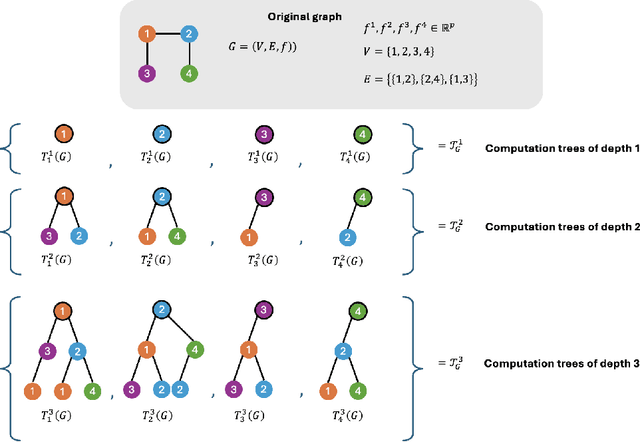
Abstract:How can we subsample graph data so that a graph neural network (GNN) trained on the subsample achieves performance comparable to training on the full dataset? This question is of fundamental interest, as smaller datasets reduce labeling costs, storage requirements, and computational resources needed for training. Selecting an effective subset is challenging: a poorly chosen subsample can severely degrade model performance, and empirically testing multiple subsets for quality obviates the benefits of subsampling. Therefore, it is critical that subsampling comes with guarantees on model performance. In this work, we introduce new subsampling methods for graph datasets that leverage the Tree Mover's Distance to reduce both the number of graphs and the size of individual graphs. To our knowledge, our approach is the first that is supported by rigorous theoretical guarantees: we prove that training a GNN on the subsampled data results in a bounded increase in loss compared to training on the full dataset. Unlike existing methods, our approach is both model-agnostic, requiring minimal assumptions about the GNN architecture, and label-agnostic, eliminating the need to label the full training set. This enables subsampling early in the model development pipeline (before data annotation, model selection, and hyperparameter tuning) reducing costs and resources needed for storage, labeling, and training. We validate our theoretical results with experiments showing that our approach outperforms existing subsampling methods across multiple datasets.
Mean-Field Sampling for Cooperative Multi-Agent Reinforcement Learning
Dec 01, 2024Abstract:Designing efficient algorithms for multi-agent reinforcement learning (MARL) is fundamentally challenging due to the fact that the size of the joint state and action spaces are exponentially large in the number of agents. These difficulties are exacerbated when balancing sequential global decision-making with local agent interactions. In this work, we propose a new algorithm \texttt{SUBSAMPLE-MFQ} (\textbf{Subsample}-\textbf{M}ean-\textbf{F}ield-\textbf{Q}-learning) and a decentralized randomized policy for a system with $n$ agents. For $k\leq n$, our algorithm system learns a policy for the system in time polynomial in $k$. We show that this learned policy converges to the optimal policy in the order of $\tilde{O}(1/\sqrt{k})$ as the number of subsampled agents $k$ increases. We validate our method empirically on Gaussian squeeze and global exploration settings.
Faster Spectral Density Estimation and Sparsification in the Nuclear Norm
Jun 11, 2024
Abstract:We consider the problem of estimating the spectral density of the normalized adjacency matrix of an $n$-node undirected graph. We provide a randomized algorithm that, with $O(n\epsilon^{-2})$ queries to a degree and neighbor oracle and in $O(n\epsilon^{-3})$ time, estimates the spectrum up to $\epsilon$ accuracy in the Wasserstein-1 metric. This improves on previous state-of-the-art methods, including an $O(n\epsilon^{-7})$ time algorithm from [Braverman et al., STOC 2022] and, for sufficiently small $\epsilon$, a $2^{O(\epsilon^{-1})}$ time method from [Cohen-Steiner et al., KDD 2018]. To achieve this result, we introduce a new notion of graph sparsification, which we call nuclear sparsification. We provide an $O(n\epsilon^{-2})$-query and $O(n\epsilon^{-2})$-time algorithm for computing $O(n\epsilon^{-2})$-sparse nuclear sparsifiers. We show that this bound is optimal in both its sparsity and query complexity, and we separate our results from the related notion of additive spectral sparsification. Of independent interest, we show that our sparsification method also yields the first deterministic algorithm for spectral density estimation that scales linearly with $n$ (sublinear in the representation size of the graph).
Truncated Variance Reduced Value Iteration
May 21, 2024Abstract:We provide faster randomized algorithms for computing an $\epsilon$-optimal policy in a discounted Markov decision process with $A_{\text{tot}}$-state-action pairs, bounded rewards, and discount factor $\gamma$. We provide an $\tilde{O}(A_{\text{tot}}[(1 - \gamma)^{-3}\epsilon^{-2} + (1 - \gamma)^{-2}])$-time algorithm in the sampling setting, where the probability transition matrix is unknown but accessible through a generative model which can be queried in $\tilde{O}(1)$-time, and an $\tilde{O}(s + (1-\gamma)^{-2})$-time algorithm in the offline setting where the probability transition matrix is known and $s$-sparse. These results improve upon the prior state-of-the-art which either ran in $\tilde{O}(A_{\text{tot}}[(1 - \gamma)^{-3}\epsilon^{-2} + (1 - \gamma)^{-3}])$ time [Sidford, Wang, Wu, Ye 2018] in the sampling setting, $\tilde{O}(s + A_{\text{tot}} (1-\gamma)^{-3})$ time [Sidford, Wang, Wu, Yang, Ye 2018] in the offline setting, or time at least quadratic in the number of states using interior point methods for linear programming. We achieve our results by building upon prior stochastic variance-reduced value iteration methods [Sidford, Wang, Wu, Yang, Ye 2018]. We provide a variant that carefully truncates the progress of its iterates to improve the variance of new variance-reduced sampling procedures that we introduce to implement the steps. Our method is essentially model-free and can be implemented in $\tilde{O}(A_{\text{tot}})$-space when given generative model access. Consequently, our results take a step in closing the sample-complexity gap between model-free and model-based methods.
From Large to Small Datasets: Size Generalization for Clustering Algorithm Selection
Feb 26, 2024



Abstract:In clustering algorithm selection, we are given a massive dataset and must efficiently select which clustering algorithm to use. We study this problem in a semi-supervised setting, with an unknown ground-truth clustering that we can only access through expensive oracle queries. Ideally, the clustering algorithm's output will be structurally close to the ground truth. We approach this problem by introducing a notion of size generalization for clustering algorithm accuracy. We identify conditions under which we can (1) subsample the massive clustering instance, (2) evaluate a set of candidate algorithms on the smaller instance, and (3) guarantee that the algorithm with the best accuracy on the small instance will have the best accuracy on the original big instance. We provide theoretical size generalization guarantees for three classic clustering algorithms: single-linkage, k-means++, and (a smoothed variant of) Gonzalez's k-centers heuristic. We validate our theoretical analysis with empirical results, observing that on real-world clustering instances, we can use a subsample of as little as 5% of the data to identify which algorithm is best on the full dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge