Hubert Banville
From Minutes to Days: Scaling Intracranial Speech Decoding with Supervised Pretraining
Dec 17, 2025Abstract:Decoding speech from brain activity has typically relied on limited neural recordings collected during short and highly controlled experiments. Here, we introduce a framework to leverage week-long intracranial and audio recordings from patients undergoing clinical monitoring, effectively increasing the training dataset size by over two orders of magnitude. With this pretraining, our contrastive learning model substantially outperforms models trained solely on classic experimental data, with gains that scale log-linearly with dataset size. Analysis of the learned representations reveals that, while brain activity represents speech features, its global structure largely drifts across days, highlighting the need for models that explicitly account for cross-day variability. Overall, our approach opens a scalable path toward decoding and modeling brain representations in both real-life and controlled task settings.
TRIBE: TRImodal Brain Encoder for whole-brain fMRI response prediction
Jul 29, 2025Abstract:Historically, neuroscience has progressed by fragmenting into specialized domains, each focusing on isolated modalities, tasks, or brain regions. While fruitful, this approach hinders the development of a unified model of cognition. Here, we introduce TRIBE, the first deep neural network trained to predict brain responses to stimuli across multiple modalities, cortical areas and individuals. By combining the pretrained representations of text, audio and video foundational models and handling their time-evolving nature with a transformer, our model can precisely model the spatial and temporal fMRI responses to videos, achieving the first place in the Algonauts 2025 brain encoding competition with a significant margin over competitors. Ablations show that while unimodal models can reliably predict their corresponding cortical networks (e.g. visual or auditory networks), they are systematically outperformed by our multimodal model in high-level associative cortices. Currently applied to perception and comprehension, our approach paves the way towards building an integrative model of representations in the human brain. Our code is available at https://github.com/facebookresearch/algonauts-2025.
Brain-to-Text Decoding: A Non-invasive Approach via Typing
Feb 18, 2025Abstract:Modern neuroprostheses can now restore communication in patients who have lost the ability to speak or move. However, these invasive devices entail risks inherent to neurosurgery. Here, we introduce a non-invasive method to decode the production of sentences from brain activity and demonstrate its efficacy in a cohort of 35 healthy volunteers. For this, we present Brain2Qwerty, a new deep learning architecture trained to decode sentences from either electro- (EEG) or magneto-encephalography (MEG), while participants typed briefly memorized sentences on a QWERTY keyboard. With MEG, Brain2Qwerty reaches, on average, a character-error-rate (CER) of 32% and substantially outperforms EEG (CER: 67%). For the best participants, the model achieves a CER of 19%, and can perfectly decode a variety of sentences outside of the training set. While error analyses suggest that decoding depends on motor processes, the analysis of typographical errors suggests that it also involves higher-level cognitive factors. Overall, these results narrow the gap between invasive and non-invasive methods and thus open the path for developing safe brain-computer interfaces for non-communicating patients.
Scaling laws for decoding images from brain activity
Jan 25, 2025



Abstract:Generative AI has recently propelled the decoding of images from brain activity. How do these approaches scale with the amount and type of neural recordings? Here, we systematically compare image decoding from four types of non-invasive devices: electroencephalography (EEG), magnetoencephalography (MEG), high-field functional Magnetic Resonance Imaging (3T fMRI) and ultra-high field (7T) fMRI. For this, we evaluate decoding models on the largest benchmark to date, encompassing 8 public datasets, 84 volunteers, 498 hours of brain recording and 2.3 million brain responses to natural images. Unlike previous work, we focus on single-trial decoding performance to simulate real-time settings. This systematic comparison reveals three main findings. First, the most precise neuroimaging devices tend to yield the best decoding performances, when the size of the training sets are similar. However, the gain enabled by deep learning - in comparison to linear models - is obtained with the noisiest devices. Second, we do not observe any plateau of decoding performance as the amount of training data increases. Rather, decoding performance scales log-linearly with the amount of brain recording. Third, this scaling law primarily depends on the amount of data per subject. However, little decoding gain is observed by increasing the number of subjects. Overall, these findings delineate the path most suitable to scale the decoding of images from non-invasive brain recordings.
Decoding individual words from non-invasive brain recordings across 723 participants
Dec 11, 2024Abstract:Deep learning has recently enabled the decoding of language from the neural activity of a few participants with electrodes implanted inside their brain. However, reliably decoding words from non-invasive recordings remains an open challenge. To tackle this issue, we introduce a novel deep learning pipeline to decode individual words from non-invasive electro- (EEG) and magneto-encephalography (MEG) signals. We train and evaluate our approach on an unprecedentedly large number of participants (723) exposed to five million words either written or spoken in English, French or Dutch. Our model outperforms existing methods consistently across participants, devices, languages, and tasks, and can decode words absent from the training set. Our analyses highlight the importance of the recording device and experimental protocol: MEG and reading are easier to decode than EEG and listening, respectively, and it is preferable to collect a large amount of data per participant than to repeat stimuli across a large number of participants. Furthermore, decoding performance consistently increases with the amount of (i) data used for training and (ii) data used for averaging during testing. Finally, single-word predictions show that our model effectively relies on word semantics but also captures syntactic and surface properties such as part-of-speech, word length and even individual letters, especially in the reading condition. Overall, our findings delineate the path and remaining challenges towards building non-invasive brain decoders for natural language.
Aligning brain functions boosts the decoding of visual semantics in novel subjects
Dec 11, 2023Abstract:Deep learning is leading to major advances in the realm of brain decoding from functional Magnetic Resonance Imaging (fMRI). However, the large inter-subject variability in brain characteristics has limited most studies to train models on one subject at a time. Consequently, this approach hampers the training of deep learning models, which typically requires very large datasets. Here, we propose to boost brain decoding by aligning brain responses to videos and static images across subjects. Compared to the anatomically-aligned baseline, our method improves out-of-subject decoding performance by up to 75%. Moreover, it also outperforms classical single-subject approaches when fewer than 100 minutes of data is available for the tested subject. Furthermore, we propose a new multi-subject alignment method, which obtains comparable results to that of classical single-subject approaches while improving out-of-subject generalization. Finally, we show that this method aligns neural representations in accordance with brain anatomy. Overall, this study lays the foundations for leveraging extensive neuroimaging datasets and enhancing the decoding of individuals with a limited amount of brain recordings.
Brain decoding: toward real-time reconstruction of visual perception
Oct 18, 2023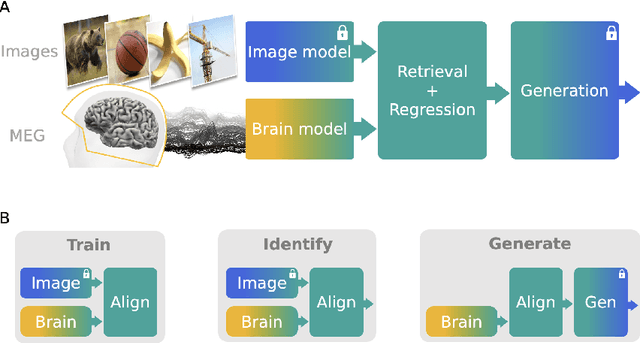
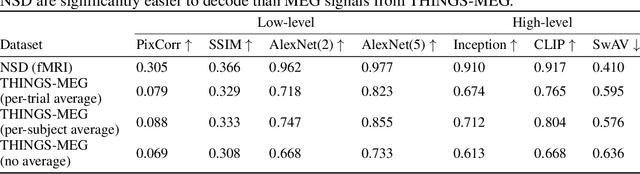
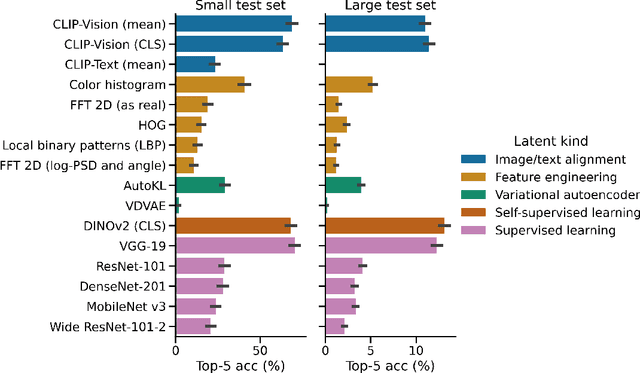
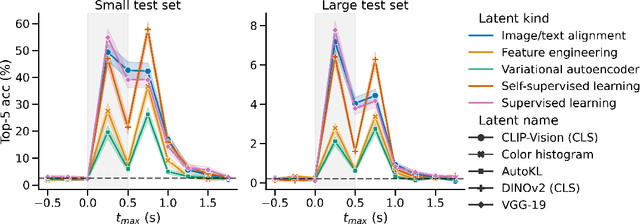
Abstract:In the past five years, the use of generative and foundational AI systems has greatly improved the decoding of brain activity. Visual perception, in particular, can now be decoded from functional Magnetic Resonance Imaging (fMRI) with remarkable fidelity. This neuroimaging technique, however, suffers from a limited temporal resolution ($\approx$0.5 Hz) and thus fundamentally constrains its real-time usage. Here, we propose an alternative approach based on magnetoencephalography (MEG), a neuroimaging device capable of measuring brain activity with high temporal resolution ($\approx$5,000 Hz). For this, we develop an MEG decoding model trained with both contrastive and regression objectives and consisting of three modules: i) pretrained embeddings obtained from the image, ii) an MEG module trained end-to-end and iii) a pretrained image generator. Our results are threefold: Firstly, our MEG decoder shows a 7X improvement of image-retrieval over classic linear decoders. Second, late brain responses to images are best decoded with DINOv2, a recent foundational image model. Third, image retrievals and generations both suggest that MEG signals primarily contain high-level visual features, whereas the same approach applied to 7T fMRI also recovers low-level features. Overall, these results provide an important step towards the decoding - in real time - of the visual processes continuously unfolding within the human brain.
Robust learning from corrupted EEG with dynamic spatial filtering
May 27, 2021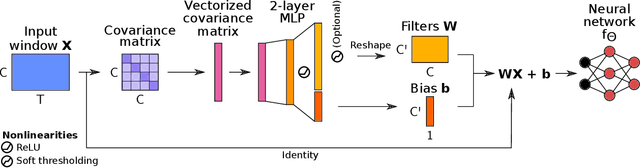
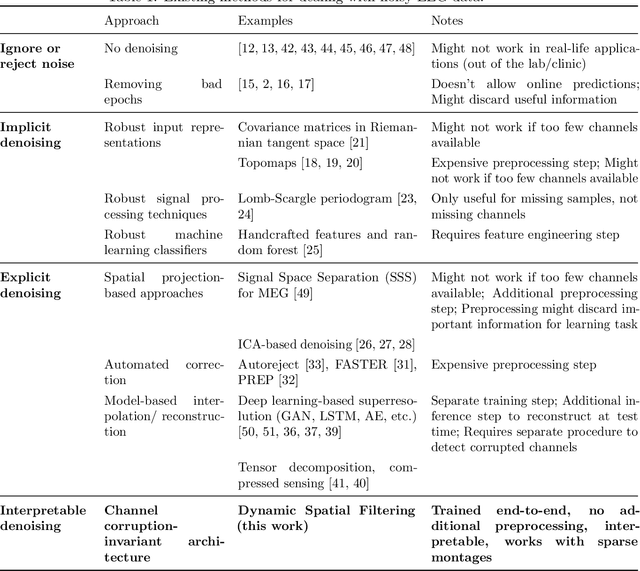
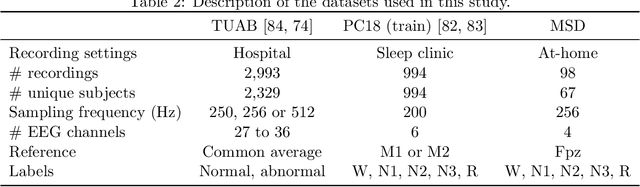
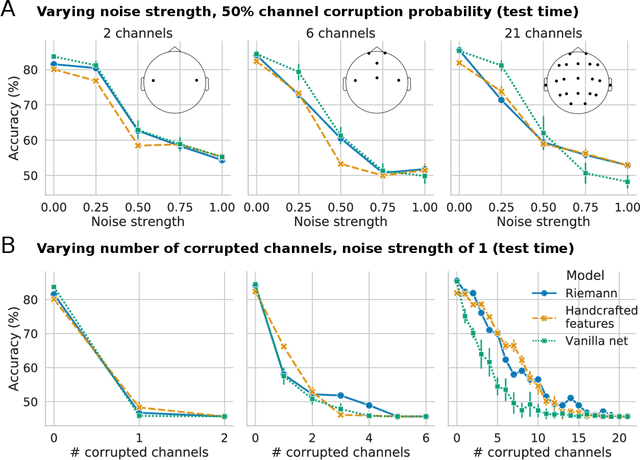
Abstract:Building machine learning models using EEG recorded outside of the laboratory setting requires methods robust to noisy data and randomly missing channels. This need is particularly great when working with sparse EEG montages (1-6 channels), often encountered in consumer-grade or mobile EEG devices. Neither classical machine learning models nor deep neural networks trained end-to-end on EEG are typically designed or tested for robustness to corruption, and especially to randomly missing channels. While some studies have proposed strategies for using data with missing channels, these approaches are not practical when sparse montages are used and computing power is limited (e.g., wearables, cell phones). To tackle this problem, we propose dynamic spatial filtering (DSF), a multi-head attention module that can be plugged in before the first layer of a neural network to handle missing EEG channels by learning to focus on good channels and to ignore bad ones. We tested DSF on public EEG data encompassing ~4,000 recordings with simulated channel corruption and on a private dataset of ~100 at-home recordings of mobile EEG with natural corruption. Our proposed approach achieves the same performance as baseline models when no noise is applied, but outperforms baselines by as much as 29.4% accuracy when significant channel corruption is present. Moreover, DSF outputs are interpretable, making it possible to monitor channel importance in real-time. This approach has the potential to enable the analysis of EEG in challenging settings where channel corruption hampers the reading of brain signals.
Uncovering the structure of clinical EEG signals with self-supervised learning
Jul 31, 2020



Abstract:Objective. Supervised learning paradigms are often limited by the amount of labeled data that is available. This phenomenon is particularly problematic in clinically-relevant data, such as electroencephalography (EEG), where labeling can be costly in terms of specialized expertise and human processing time. Consequently, deep learning architectures designed to learn on EEG data have yielded relatively shallow models and performances at best similar to those of traditional feature-based approaches. However, in most situations, unlabeled data is available in abundance. By extracting information from this unlabeled data, it might be possible to reach competitive performance with deep neural networks despite limited access to labels. Approach. We investigated self-supervised learning (SSL), a promising technique for discovering structure in unlabeled data, to learn representations of EEG signals. Specifically, we explored two tasks based on temporal context prediction as well as contrastive predictive coding on two clinically-relevant problems: EEG-based sleep staging and pathology detection. We conducted experiments on two large public datasets with thousands of recordings and performed baseline comparisons with purely supervised and hand-engineered approaches. Main results. Linear classifiers trained on SSL-learned features consistently outperformed purely supervised deep neural networks in low-labeled data regimes while reaching competitive performance when all labels were available. Additionally, the embeddings learned with each method revealed clear latent structures related to physiological and clinical phenomena, such as age effects. Significance. We demonstrate the benefit of self-supervised learning approaches on EEG data. Our results suggest that SSL may pave the way to a wider use of deep learning models on EEG data.
Self-supervised representation learning from electroencephalography signals
Nov 13, 2019



Abstract:The supervised learning paradigm is limited by the cost - and sometimes the impracticality - of data collection and labeling in multiple domains. Self-supervised learning, a paradigm which exploits the structure of unlabeled data to create learning problems that can be solved with standard supervised approaches, has shown great promise as a pretraining or feature learning approach in fields like computer vision and time series processing. In this work, we present self-supervision strategies that can be used to learn informative representations from multivariate time series. One successful approach relies on predicting whether time windows are sampled from the same temporal context or not. As demonstrated on a clinically relevant task (sleep scoring) and with two electroencephalography datasets, our approach outperforms a purely supervised approach in low data regimes, while capturing important physiological information without any access to labels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge