Hans Liebl
Department of Diagnostic and Interventional Neuroradiology, School of Medicine, Klinikum rechts der Isar, Technical University of Munich, Germany
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
A Computed Tomography Vertebral Segmentation Dataset with Anatomical Variations and Multi-Vendor Scanner Data
Mar 10, 2021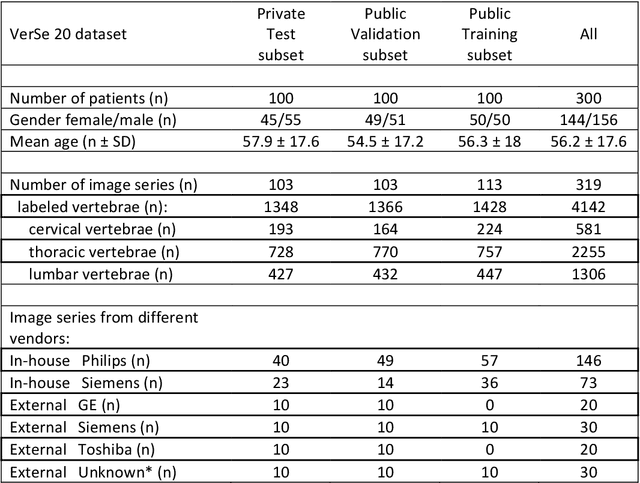
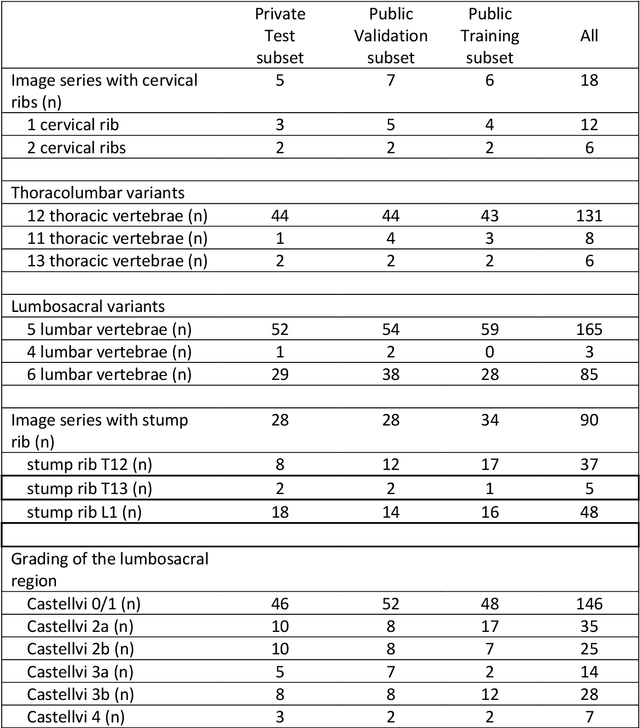
Abstract:With the advent of deep learning algorithms, fully automated radiological image analysis is within reach. In spine imaging, several atlas- and shape-based as well as deep learning segmentation algorithms have been proposed, allowing for subsequent automated analysis of morphology and pathology. The first Large Scale Vertebrae Segmentation Challenge (VerSe 2019) showed that these perform well on normal anatomy, but fail in variants not frequently present in the training dataset. Building on that experience, we report on the largely increased VerSe 2020 dataset and results from the second iteration of the VerSe challenge (MICCAI 2020, Lima, Peru). VerSe 2020 comprises annotated spine computed tomography (CT) images from 300 subjects with 4142 fully visualized and annotated vertebrae, collected across multiple centres from four different scanner manufacturers, enriched with cases that exhibit anatomical variants such as enumeration abnormalities (n=77) and transitional vertebrae (n=161). Metadata includes vertebral labelling information, voxel-level segmentation masks obtained with a human-machine hybrid algorithm and anatomical ratings, to enable the development and benchmarking of robust and accurate segmentation algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge