Haijian Chen
Improving Masked Autoencoders by Learning Where to Mask
Mar 12, 2023


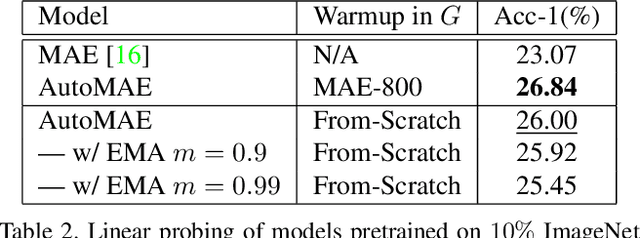
Abstract:Masked image modeling is a promising self-supervised learning method for visual data. It is typically built upon image patches with random masks, which largely ignores the variation of information density between them. The question is: Is there a better masking strategy than random sampling and how can we learn it? We empirically study this problem and initially find that introducing object-centric priors in mask sampling can significantly improve the learned representations. Inspired by this observation, we present AutoMAE, a fully differentiable framework that uses Gumbel-Softmax to interlink an adversarially-trained mask generator and a mask-guided image modeling process. In this way, our approach can adaptively find patches with higher information density for different images, and further strike a balance between the information gain obtained from image reconstruction and its practical training difficulty. In our experiments, AutoMAE is shown to provide effective pretraining models on standard self-supervised benchmarks and downstream tasks.
A translational pathway of deep learning methods in GastroIntestinal Endoscopy
Oct 12, 2020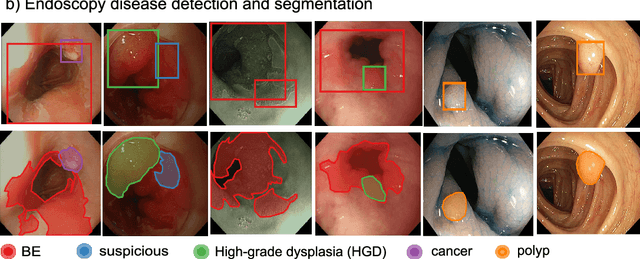
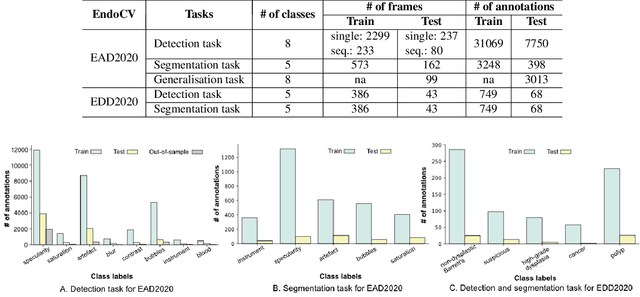
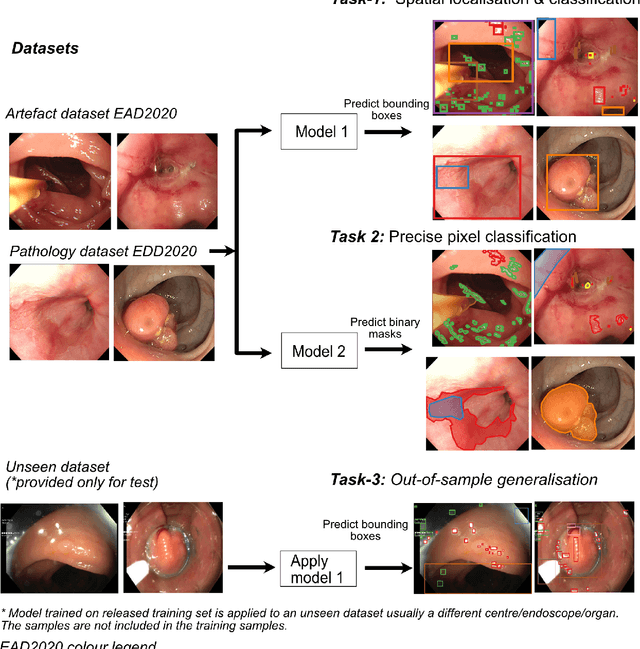
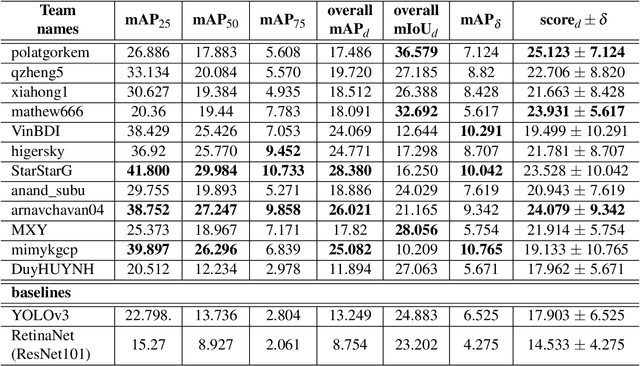
Abstract:The Endoscopy Computer Vision Challenge (EndoCV) is a crowd-sourcing initiative to address eminent problems in developing reliable computer aided detection and diagnosis endoscopy systems and suggest a pathway for clinical translation of technologies. Whilst endoscopy is a widely used diagnostic and treatment tool for hollow-organs, there are several core challenges often faced by endoscopists, mainly: 1) presence of multi-class artefacts that hinder their visual interpretation, and 2) difficulty in identifying subtle precancerous precursors and cancer abnormalities. Artefacts often affect the robustness of deep learning methods applied to the gastrointestinal tract organs as they can be confused with tissue of interest. EndoCV2020 challenges are designed to address research questions in these remits. In this paper, we present a summary of methods developed by the top 17 teams and provide an objective comparison of state-of-the-art methods and methods designed by the participants for two sub-challenges: i) artefact detection and segmentation (EAD2020), and ii) disease detection and segmentation (EDD2020). Multi-center, multi-organ, multi-class, and multi-modal clinical endoscopy datasets were compiled for both EAD2020 and EDD2020 sub-challenges. An out-of-sample generalisation ability of detection algorithms was also evaluated. Whilst most teams focused on accuracy improvements, only a few methods hold credibility for clinical usability. The best performing teams provided solutions to tackle class imbalance, and variabilities in size, origin, modality and occurrences by exploring data augmentation, data fusion, and optimal class thresholding techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge