Hady Ahmady Phoulady
The Weighted Tsetlin Machine: Compressed Representations with Weighted Clauses
Jan 14, 2020


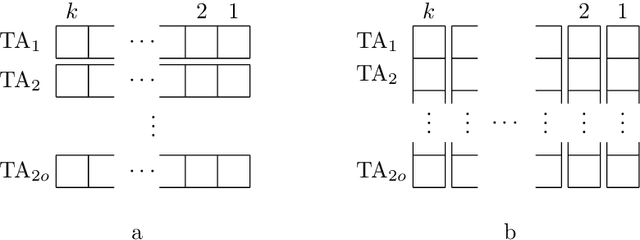
Abstract:The Tsetlin Machine (TM) is an interpretable mechanism for pattern recognition that constructs conjunctive clauses from data. The clauses capture frequent patterns with high discriminating power, providing increasing expression power with each additional clause. However, the resulting accuracy gain comes at the cost of linear growth in computation time and memory usage. In this paper, we present the Weighted Tsetlin Machine (WTM), which reduces computation time and memory usage by weighting the clauses. Real-valued weighting allows one clause to replace multiple, and supports fine-tuning the impact of each clause. Our novel scheme simultaneously learns both the composition of the clauses and their weights. Furthermore, we increase training efficiency by replacing $k$ Bernoulli trials of success probability $p$ with a uniform sample of average size $p k$, the size drawn from a binomial distribution. In our empirical evaluation, the WTM achieved the same accuracy as the TM on MNIST, IMDb, and Connect-4, requiring only $1/4$, $1/3$, and $1/50$ of the clauses, respectively. With the same number of clauses, the WTM outperformed the TM, obtaining peak test accuracies of respectively $98.63\%$, $90.37\%$, and $87.91\%$. Finally, our novel sampling scheme reduced sample generation time by a factor of $7$.
Iterative Deep Learning Based Unbiased Stereology With Human-in-the-Loop
Jan 14, 2019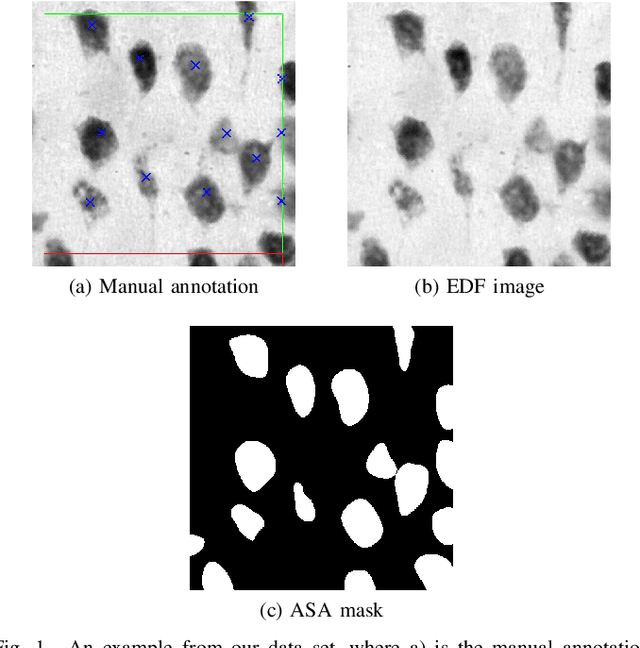
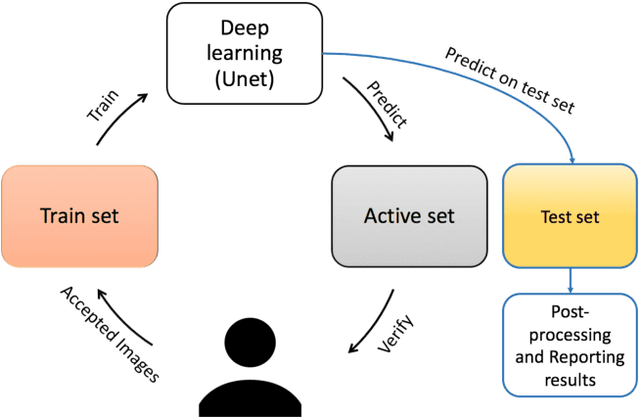
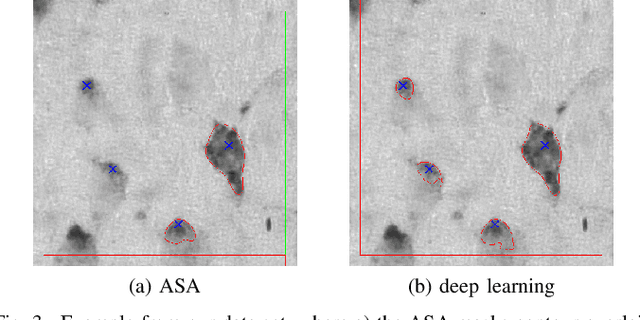
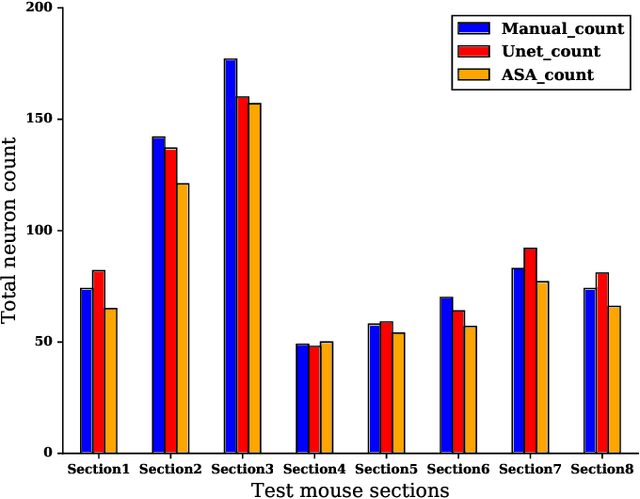
Abstract:Lack of enough labeled data is a major problem in building machine learning based models when the manual annotation (labeling) is error-prone, expensive, tedious, and time-consuming. In this paper, we introduce an iterative deep learning based method to improve segmentation and counting of cells based on unbiased stereology applied to regions of interest of extended depth of field (EDF) images. This method uses an existing machine learning algorithm called the adaptive segmentation algorithm (ASA) to generate masks (verified by a user) for EDF images to train deep learning models. Then an iterative deep learning approach is used to feed newly predicted and accepted deep learning masks/images (verified by a user) to the training set of the deep learning model. The error rate in unbiased stereology count of cells on an unseen test set reduced from about 3 % to less than 1 % after 5 iterations of the iterative deep learning based unbiased stereology process.
A New Cervical Cytology Dataset for Nucleus Detection and Image Classification and Methods for Cervical Nucleus Detection
Nov 23, 2018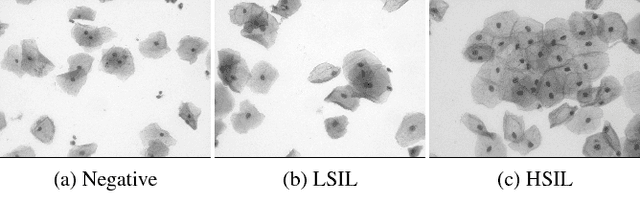

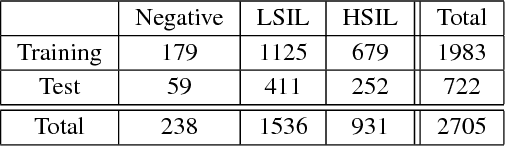
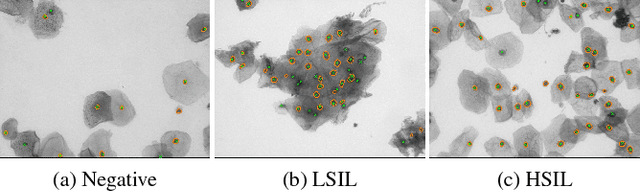
Abstract:Analyzing Pap cytology slides is an important tasks in detecting and grading precancerous and cancerous cervical cancer stages. Processing cytology images usually involve segmenting nuclei and overlapping cells. We introduce a cervical cytology dataset that can be used to evaluate nucleus detection, as well as image classification methods in the cytology image processing area. This dataset contains 93 real image stacks with their grade labels and manually annotated nuclei within images. We also present two methods: a baseline method based on a previously proposed approach, and a deep learning method, and compare their results with other state-of-the-art methods. Both the baseline method and the deep learning method outperform other state-of-the-art methods by significant margins. Along with the dataset, we publicly make the evaluation code and the baseline method available to download for further benchmarking.
Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge
Jul 22, 2018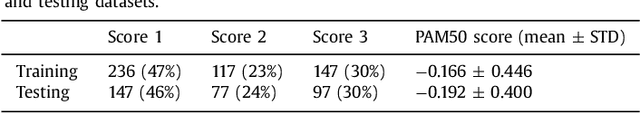
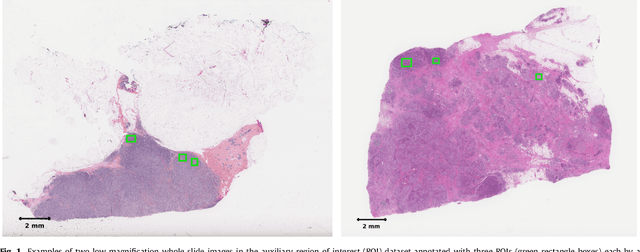
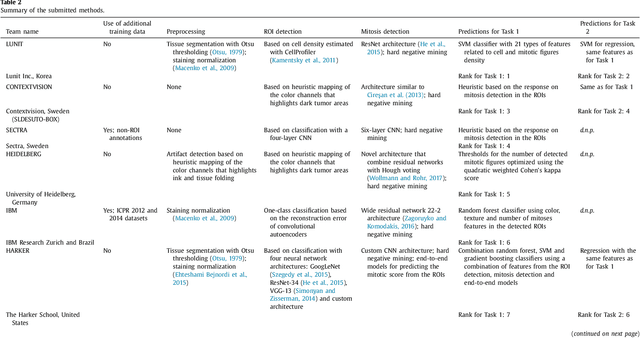
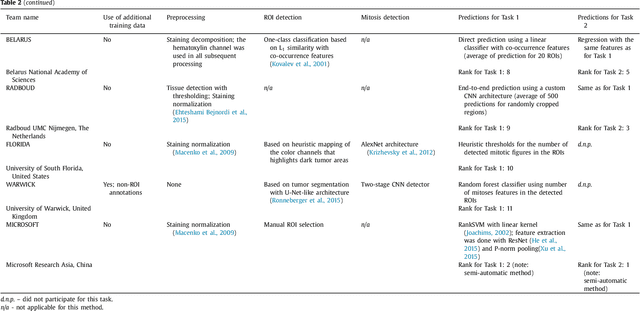
Abstract:Tumor proliferation is an important biomarker indicative of the prognosis of breast cancer patients. Assessment of tumor proliferation in a clinical setting is highly subjective and labor-intensive task. Previous efforts to automate tumor proliferation assessment by image analysis only focused on mitosis detection in predefined tumor regions. However, in a real-world scenario, automatic mitosis detection should be performed in whole-slide images (WSIs) and an automatic method should be able to produce a tumor proliferation score given a WSI as input. To address this, we organized the TUmor Proliferation Assessment Challenge 2016 (TUPAC16) on prediction of tumor proliferation scores from WSIs. The challenge dataset consisted of 500 training and 321 testing breast cancer histopathology WSIs. In order to ensure fair and independent evaluation, only the ground truth for the training dataset was provided to the challenge participants. The first task of the challenge was to predict mitotic scores, i.e., to reproduce the manual method of assessing tumor proliferation by a pathologist. The second task was to predict the gene expression based PAM50 proliferation scores from the WSI. The best performing automatic method for the first task achieved a quadratic-weighted Cohen's kappa score of $\kappa$ = 0.567, 95% CI [0.464, 0.671] between the predicted scores and the ground truth. For the second task, the predictions of the top method had a Spearman's correlation coefficient of r = 0.617, 95% CI [0.581 0.651] with the ground truth. This was the first study that investigated tumor proliferation assessment from WSIs. The achieved results are promising given the difficulty of the tasks and weakly-labelled nature of the ground truth. However, further research is needed to improve the practical utility of image analysis methods for this task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge