Karl Rohr
CellCentroidFormer: Combining Self-attention and Convolution for Cell Detection
Jun 01, 2022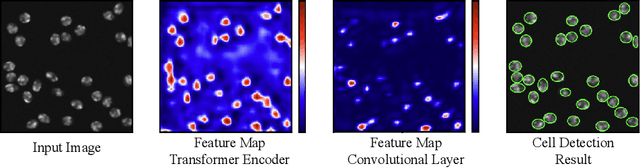
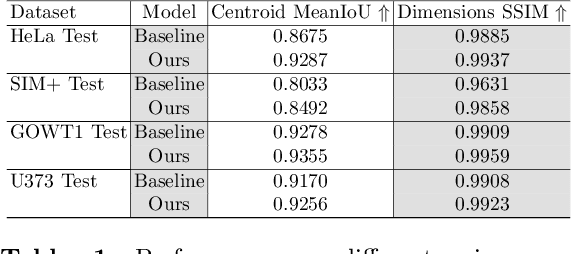
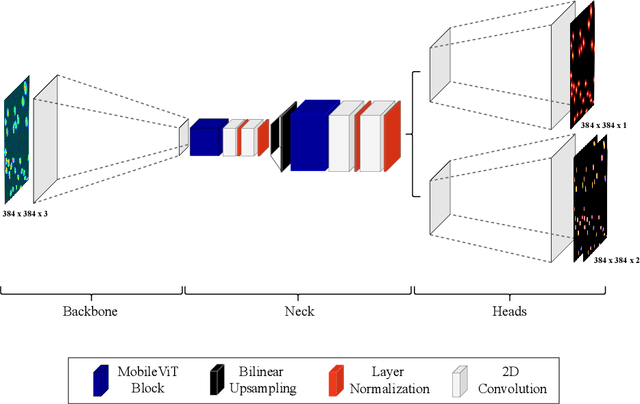

Abstract:Cell detection in microscopy images is important to study how cells move and interact with their environment. Most recent deep learning-based methods for cell detection use convolutional neural networks (CNNs). However, inspired by the success in other computer vision applications, vision transformers (ViTs) are also used for this purpose. We propose a novel hybrid CNN-ViT model for cell detection in microscopy images to exploit the advantages of both types of deep learning models. We employ an efficient CNN, that was pre-trained on the ImageNet dataset, to extract image features and utilize transfer learning to reduce the amount of required training data. Extracted image features are further processed by a combination of convolutional and transformer layers, so that the convolutional layers can focus on local information and the transformer layers on global information. Our centroid-based cell detection method represents cells as ellipses and is end-to-end trainable. Furthermore, we show that our proposed model can outperform a fully convolutional baseline model on four different 2D microscopy datasets. Code is available at: https://github.com/roydenwa/cell-centroid-former
EfficientCellSeg: Efficient Volumetric Cell Segmentation Using Context Aware Pseudocoloring
Apr 06, 2022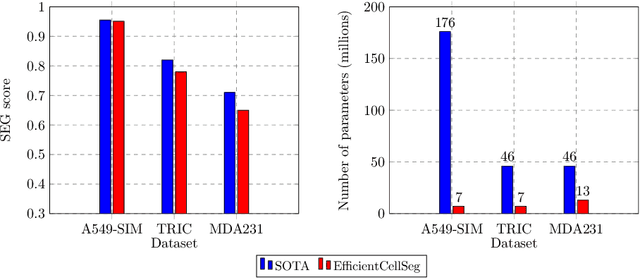

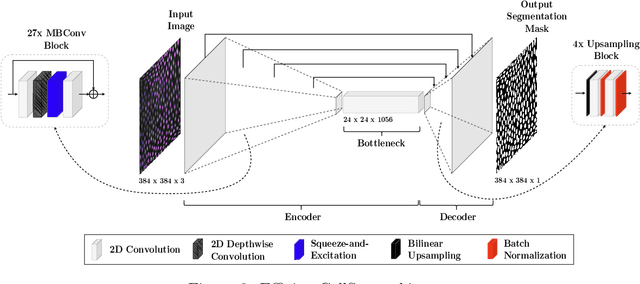

Abstract:Volumetric cell segmentation in fluorescence microscopy images is important to study a wide variety of cellular processes. Applications range from the analysis of cancer cells to behavioral studies of cells in the embryonic stage. Like in other computer vision fields, most recent methods use either large convolutional neural networks (CNNs) or vision transformer models (ViTs). Since the number of available 3D microscopy images is typically limited in applications, we take a different approach and introduce a small CNN for volumetric cell segmentation. Compared to previous CNN models for cell segmentation, our model is efficient and has an asymmetric encoder-decoder structure with very few parameters in the decoder. Training efficiency is further improved via transfer learning. In addition, we introduce Context Aware Pseudocoloring to exploit spatial context in z-direction of 3D images while performing volumetric cell segmentation slice-wise. We evaluated our method using different 3D datasets from the Cell Segmentation Benchmark of the Cell Tracking Challenge. Our segmentation method achieves top-ranking results, while our CNN model has an up to 25x lower number of parameters than other top-ranking methods. Code and pretrained models are available at: https://github.com/roydenwa/efficient-cell-seg
Towards ultra-high resolution 3D reconstruction of a whole rat brain from 3D-PLI data
Jul 29, 2018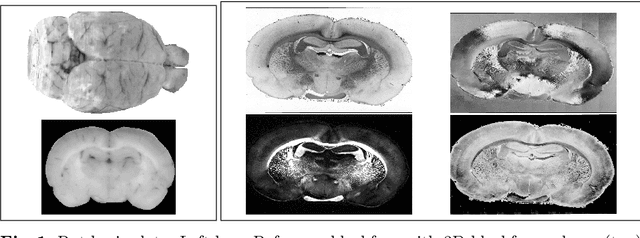

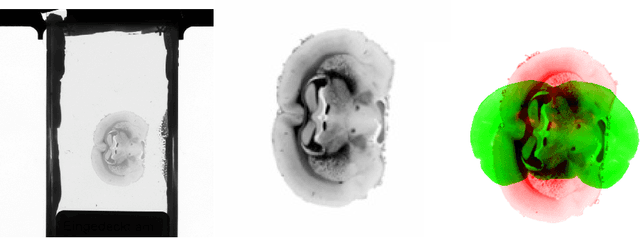

Abstract:3D reconstruction of the fiber connectivity of the rat brain at microscopic scale enables gaining detailed insight about the complex structural organization of the brain. We introduce a new method for registration and 3D reconstruction of high- and ultra-high resolution (64 $\mu$m and 1.3 $\mu$m pixel size) histological images of a Wistar rat brain acquired by 3D polarized light imaging (3D-PLI). Our method exploits multi-scale and multi-modal 3D-PLI data up to cellular resolution. We propose a new feature transform-based similarity measure and a weighted regularization scheme for accurate and robust non-rigid registration. To transform the 1.3 $\mu$m ultra-high resolution data to the reference blockface images a feature-based registration method followed by a non-rigid registration is proposed. Our approach has been successfully applied to 278 histological sections of a rat brain and the performance has been quantitatively evaluated using manually placed landmarks by an expert.
Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge
Jul 22, 2018
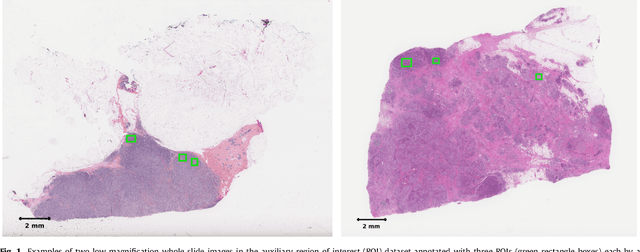
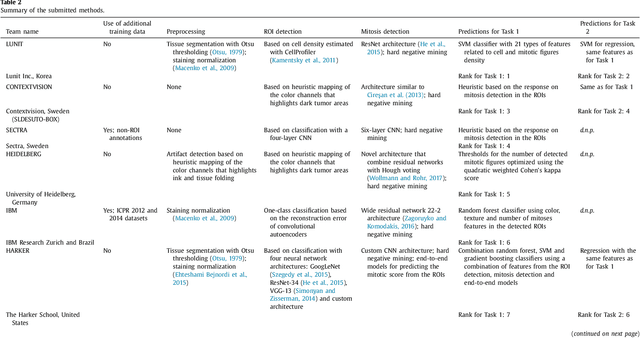
Abstract:Tumor proliferation is an important biomarker indicative of the prognosis of breast cancer patients. Assessment of tumor proliferation in a clinical setting is highly subjective and labor-intensive task. Previous efforts to automate tumor proliferation assessment by image analysis only focused on mitosis detection in predefined tumor regions. However, in a real-world scenario, automatic mitosis detection should be performed in whole-slide images (WSIs) and an automatic method should be able to produce a tumor proliferation score given a WSI as input. To address this, we organized the TUmor Proliferation Assessment Challenge 2016 (TUPAC16) on prediction of tumor proliferation scores from WSIs. The challenge dataset consisted of 500 training and 321 testing breast cancer histopathology WSIs. In order to ensure fair and independent evaluation, only the ground truth for the training dataset was provided to the challenge participants. The first task of the challenge was to predict mitotic scores, i.e., to reproduce the manual method of assessing tumor proliferation by a pathologist. The second task was to predict the gene expression based PAM50 proliferation scores from the WSI. The best performing automatic method for the first task achieved a quadratic-weighted Cohen's kappa score of $\kappa$ = 0.567, 95% CI [0.464, 0.671] between the predicted scores and the ground truth. For the second task, the predictions of the top method had a Spearman's correlation coefficient of r = 0.617, 95% CI [0.581 0.651] with the ground truth. This was the first study that investigated tumor proliferation assessment from WSIs. The achieved results are promising given the difficulty of the tasks and weakly-labelled nature of the ground truth. However, further research is needed to improve the practical utility of image analysis methods for this task.
Automatic breast cancer grading in lymph nodes using a deep neural network
Jul 24, 2017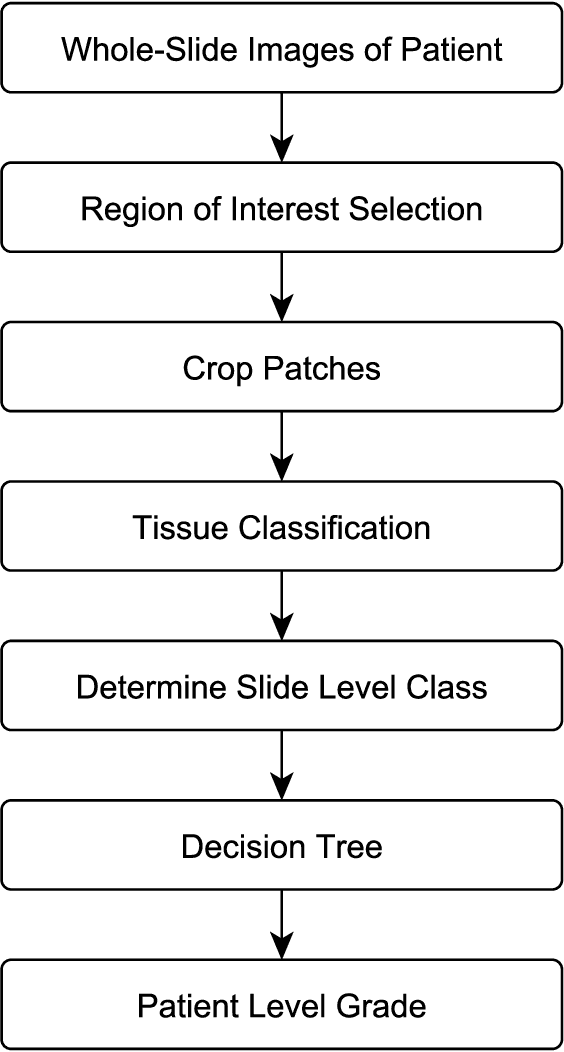

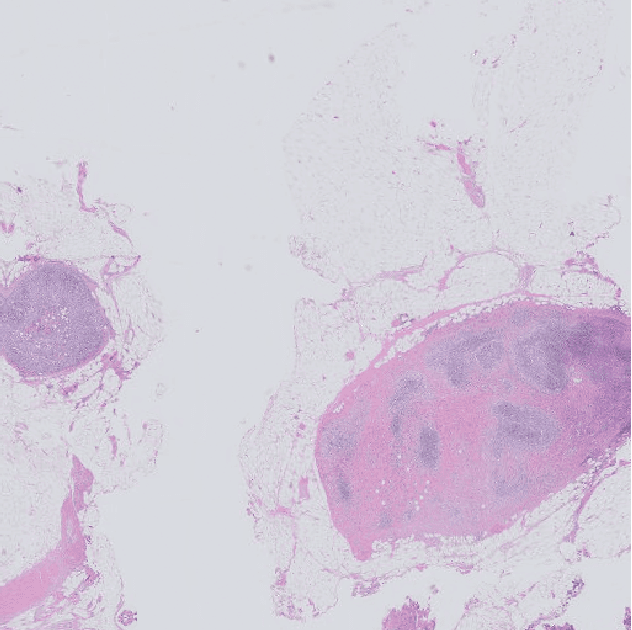
Abstract:The progression of breast cancer can be quantified in lymph node whole-slide images (WSIs). We describe a novel method for effectively performing classification of whole-slide images and patient level breast cancer grading. Our method utilises a deep neural network. The method performs classification on small patches and uses model averaging for boosting. In the first step, region of interest patches are determined and cropped automatically by color thresholding and then classified by the deep neural network. The classification results are used to determine a slide level class and for further aggregation to predict a patient level grade. Fast processing speed of our method enables high throughput image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge