EfficientCellSeg: Efficient Volumetric Cell Segmentation Using Context Aware Pseudocoloring
Paper and Code
Apr 06, 2022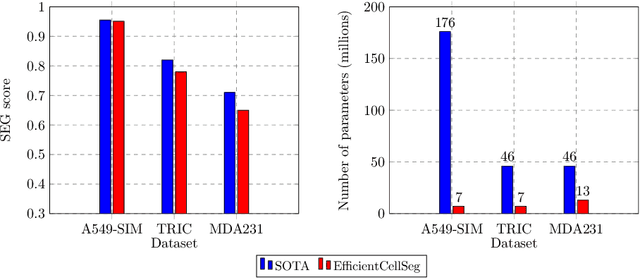

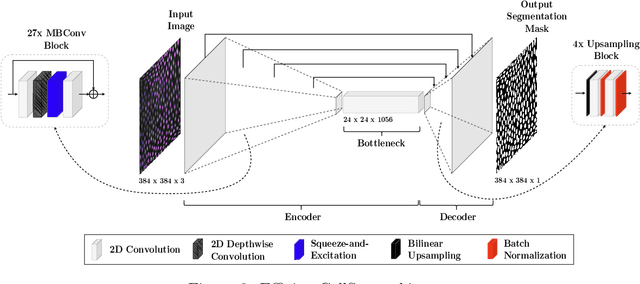

Volumetric cell segmentation in fluorescence microscopy images is important to study a wide variety of cellular processes. Applications range from the analysis of cancer cells to behavioral studies of cells in the embryonic stage. Like in other computer vision fields, most recent methods use either large convolutional neural networks (CNNs) or vision transformer models (ViTs). Since the number of available 3D microscopy images is typically limited in applications, we take a different approach and introduce a small CNN for volumetric cell segmentation. Compared to previous CNN models for cell segmentation, our model is efficient and has an asymmetric encoder-decoder structure with very few parameters in the decoder. Training efficiency is further improved via transfer learning. In addition, we introduce Context Aware Pseudocoloring to exploit spatial context in z-direction of 3D images while performing volumetric cell segmentation slice-wise. We evaluated our method using different 3D datasets from the Cell Segmentation Benchmark of the Cell Tracking Challenge. Our segmentation method achieves top-ranking results, while our CNN model has an up to 25x lower number of parameters than other top-ranking methods. Code and pretrained models are available at: https://github.com/roydenwa/efficient-cell-seg
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge