Lawrence O. Hall
Reducing Data Requirements for Sequence-Property Prediction in Copolymer Compatibilizers via Deep Neural Network Tuning
Jul 29, 2025Abstract:Synthetic sequence-controlled polymers promise to transform polymer science by combining the chemical versatility of synthetic polymers with the precise sequence-mediated functionality of biological proteins. However, design of these materials has proven extraordinarily challenging, because they lack the massive datasets of closely related evolved molecules that accelerate design of proteins. Here we report on a new Artifical Intelligence strategy to dramatically reduce the amount of data necessary to accelerate these materials' design. We focus on data connecting the repeat-unit-sequence of a \emph{compatibilizer} molecule to its ability to reduce the interfacial tension between distinct polymer domains. The optimal sequence of these molecules, which are essential for applications such as mixed-waste polymer recycling, depends strongly on variables such as concentration and chemical details of the polymer. With current methods, this would demand an entirely distinct dataset to enable design at each condition. Here we show that a deep neural network trained on low-fidelity data for sequence/interfacial tension relations at one set of conditions can be rapidly tuned to make higher-fidelity predictions at a distinct set of conditions, requiring far less data that would ordinarily be needed. This priming-and-tuning approach should allow a single low-fidelity parent dataset to dramatically accelerate prediction and design in an entire constellation of related systems. In the long run, it may also provide an approach to bootstrapping quantitative atomistic design with AI insights from fast, coarse simulations.
Active Prompt Tuning Enables Gpt-40 To Do Efficient Classification Of Microscopy Images
Nov 04, 2024Abstract:Traditional deep learning-based methods for classifying cellular features in microscopy images require time- and labor-intensive processes for training models. Among the current limitations are major time commitments from domain experts for accurate ground truth preparation; and the need for a large amount of input image data. We previously proposed a solution that overcomes these challenges using OpenAI's GPT-4(V) model on a pilot dataset (Iba-1 immuno-stained tissue sections from 11 mouse brains). Results on the pilot dataset were equivalent in accuracy and with a substantial improvement in throughput efficiency compared to the baseline using a traditional Convolutional Neural Net (CNN)-based approach. The present study builds upon this framework using a second unique and substantially larger dataset of microscopy images. Our current approach uses a newer and faster model, GPT-4o, along with improved prompts. It was evaluated on a microscopy image dataset captured at low (10x) magnification from cresyl-violet-stained sections through the cerebellum of a total of 18 mouse brains (9 Lurcher mice, 9 wild-type controls). We used our approach to classify these images either as a control group or Lurcher mutant. Using 6 mice in the prompt set the results were correct classification for 11 out of the 12 mice (92%) with 96% higher efficiency, reduced image requirements, and lower demands on time and effort of domain experts compared to the baseline method (snapshot ensemble of CNN models). These results confirm that our approach is effective across multiple datasets from different brain regions and magnifications, with minimal overhead.
Analysis of MRI Biomarkers for Brain Cancer Survival Prediction
Sep 03, 2021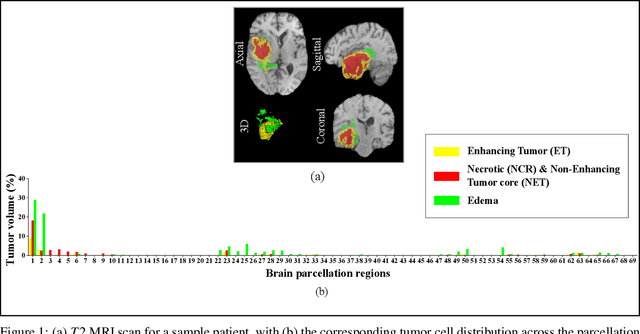
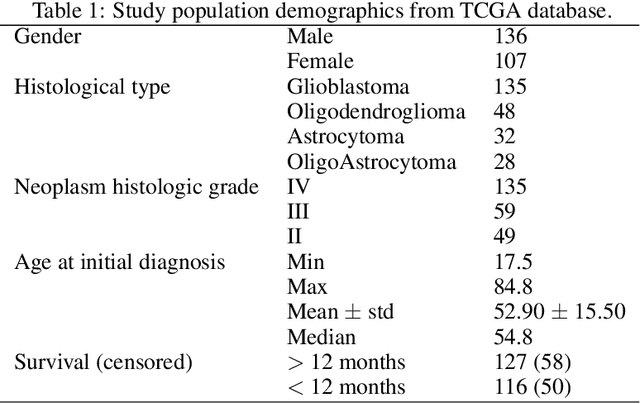
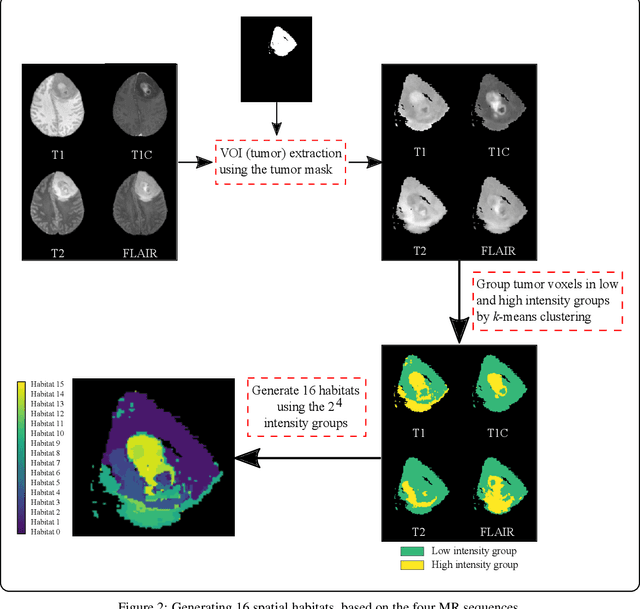
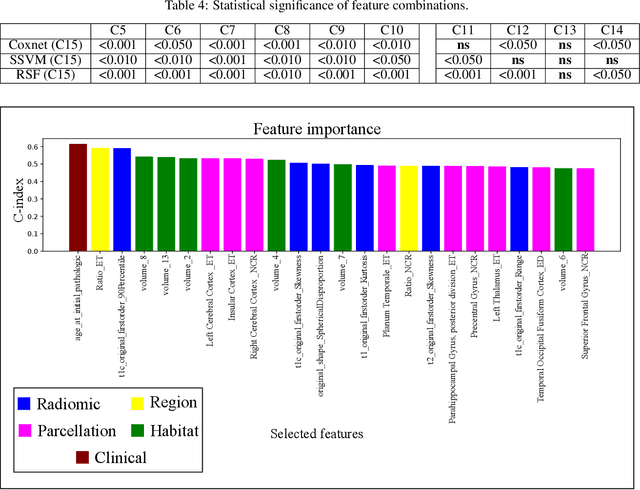
Abstract:Prediction of Overall Survival (OS) of brain cancer patients from multi-modal MRI is a challenging field of research. Most of the existing literature on survival prediction is based on Radiomic features, which does not consider either non-biological factors or the functional neurological status of the patient(s). Besides, the selection of an appropriate cut-off for survival and the presence of censored data create further problems. Application of deep learning models for OS prediction is also limited due to the lack of large annotated publicly available datasets. In this scenario we analyse the potential of two novel neuroimaging feature families, extracted from brain parcellation atlases and spatial habitats, along with classical radiomic and geometric features; to study their combined predictive power for analysing overall survival. A cross validation strategy with grid search is proposed to simultaneously select and evaluate the most predictive feature subset based on its predictive power. A Cox Proportional Hazard (CoxPH) model is employed for univariate feature selection, followed by the prediction of patient-specific survival functions by three multivariate parsimonious models viz. Coxnet, Random survival forests (RSF) and Survival SVM (SSVM). The brain cancer MRI data used for this research was taken from two open-access collections TCGA-GBM and TCGA-LGG available from The Cancer Imaging Archive (TCIA). Corresponding survival data for each patient was downloaded from The Cancer Genome Atlas (TCGA). A high cross validation $C-index$ score of $0.82\pm.10$ was achieved using RSF with the best $24$ selected features. Age was found to be the most important biological predictor. There were $9$, $6$, $6$ and $2$ features selected from the parcellation, habitat, radiomic and region-based feature groups respectively.
Deep Learning Models May Spuriously Classify Covid-19 from X-ray Images Based on Confounders
Jan 08, 2021
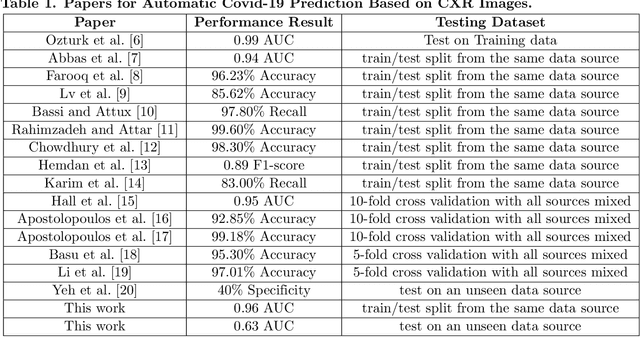
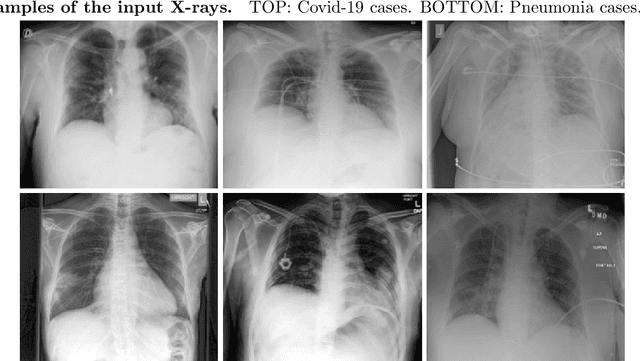
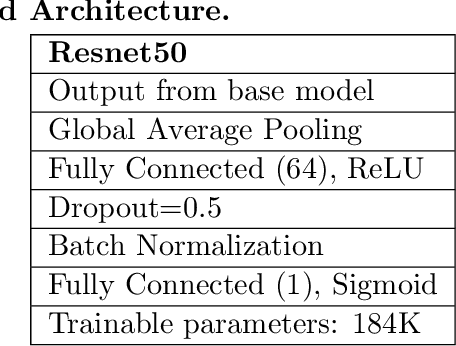
Abstract:Identifying who is infected with the Covid-19 virus is critical for controlling its spread. X-ray machines are widely available worldwide and can quickly provide images that can be used for diagnosis. A number of recent studies claim it may be possible to build highly accurate models, using deep learning, to detect Covid-19 from chest X-ray images. This paper explores the robustness and generalization ability of convolutional neural network models in diagnosing Covid-19 disease from frontal-view (AP/PA), raw chest X-ray images that were lung field cropped. Some concerning observations are made about high performing models that have learned to rely on confounding features related to the data source, rather than the patient's lung pathology, when differentiating between Covid-19 positive and negative labels. Specifically, these models likely made diagnoses based on confounding factors such as patient age or image processing artifacts, rather than medically relevant information.
Finding Covid-19 from Chest X-rays using Deep Learning on a Small Dataset
Apr 13, 2020Abstract:Testing for COVID-19 has been unable to keep up with the demand. Further, the false negative rate is projected to be as high as 30% and test results can take some time to obtain. X-ray machines are widely available and provide images for diagnosis quickly. This paper explores how useful chest X-ray images can be in diagnosing COVID-19 disease. We have obtained 122 chest X-rays of COVID-19 and over 4,000 chest X-rays of viral and bacterial pneumonia. A pretrained deep convolutional neural network has been tuned on 102 COVID-19 cases and 102 other pneumonia cases in a 10-fold cross validation. The results were all 102 COVID-19 cases were correctly classified and there were 8 false positives resulting in an AUC of 0.997. On a test set of 20 unseen COVID-19 cases all were correctly classified and more than 95% of 4171 other pneumonia examples were correctly classified. This study has flaws, most critically a lack of information about where in the disease process the COVID-19 cases were and the small data set size. More COVID-19 case images will enable a better answer to the question of how useful chest X-rays can be for diagnosing COVID-19 (so please send them).
Iterative Deep Learning Based Unbiased Stereology With Human-in-the-Loop
Jan 14, 2019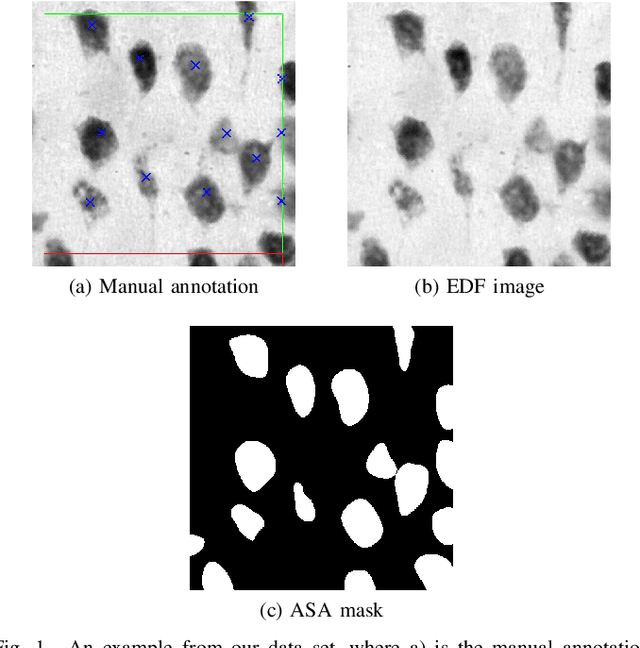
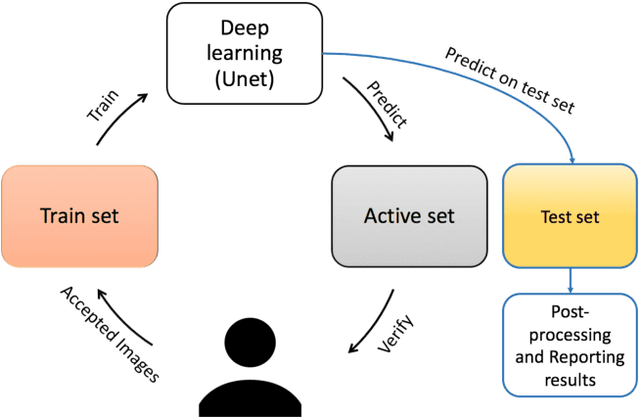
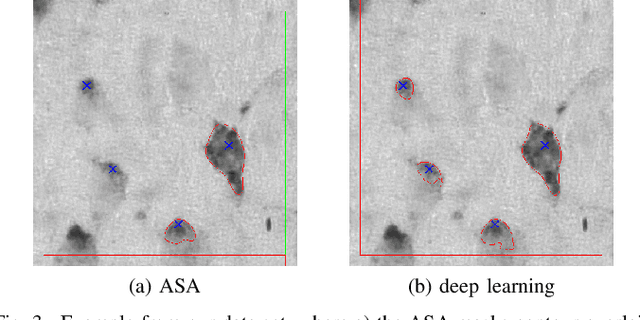
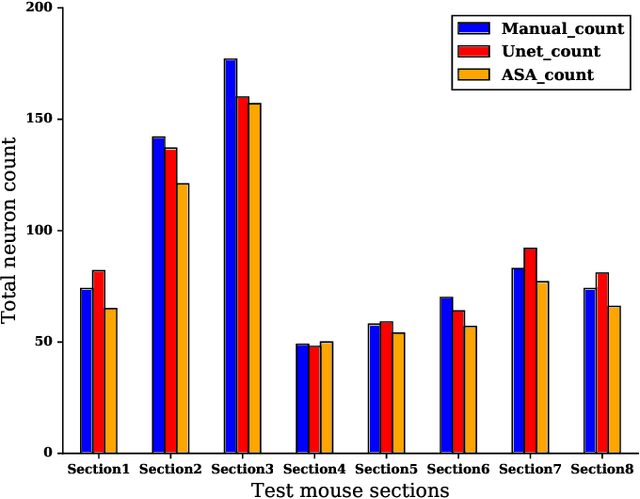
Abstract:Lack of enough labeled data is a major problem in building machine learning based models when the manual annotation (labeling) is error-prone, expensive, tedious, and time-consuming. In this paper, we introduce an iterative deep learning based method to improve segmentation and counting of cells based on unbiased stereology applied to regions of interest of extended depth of field (EDF) images. This method uses an existing machine learning algorithm called the adaptive segmentation algorithm (ASA) to generate masks (verified by a user) for EDF images to train deep learning models. Then an iterative deep learning approach is used to feed newly predicted and accepted deep learning masks/images (verified by a user) to the training set of the deep learning model. The error rate in unbiased stereology count of cells on an unseen test set reduced from about 3 % to less than 1 % after 5 iterations of the iterative deep learning based unbiased stereology process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge