Sushmita Mitra
Efficient Self-Supervised Grading of Prostate Cancer Pathology
Jan 26, 2025Abstract:Prostate cancer grading using the ISUP system (International Society of Urological Pathology) for treatment decisions is highly subjective and requires considerable expertise. Despite advances in computer-aided diagnosis systems, few have handled efficient ISUP grading on Whole Slide Images (WSIs) of prostate biopsies based only on slide-level labels. Some of the general challenges include managing gigapixel WSIs, obtaining patch-level annotations, and dealing with stain variability across centers. One of the main task-specific challenges faced by deep learning in ISUP grading, is the learning of patch-level features of Gleason patterns (GPs) based only on their slide labels. In this scenario, an efficient framework for ISUP grading is developed. The proposed TSOR is based on a novel Task-specific Self-supervised learning (SSL) model, which is fine-tuned using Ordinal Regression. Since the diversity of training samples plays a crucial role in SSL, a patch-level dataset is created to be relatively balanced w.r.t. the Gleason grades (GGs). This balanced dataset is used for pre-training, so that the model can effectively learn stain-agnostic features of the GP for better generalization. In medical image grading, it is desirable that misclassifications be as close as possible to the actual grade. From this perspective, the model is then fine-tuned for the task of ISUP grading using an ordinal regression-based approach. Experimental results on the most extensive multicenter prostate biopsies dataset (PANDA challenge), as well as the SICAP dataset, demonstrate the effectiveness of this novel framework compared to state-of-the-art methods.
Adaptive Class Learning to Screen Diabetic Disorders in Fundus Images of Eye
Jan 21, 2025Abstract:The prevalence of ocular illnesses is growing globally, presenting a substantial public health challenge. Early detection and timely intervention are crucial for averting visual impairment and enhancing patient prognosis. This research introduces a new framework called Class Extension with Limited Data (CELD) to train a classifier to categorize retinal fundus images. The classifier is initially trained to identify relevant features concerning Healthy and Diabetic Retinopathy (DR) classes and later fine-tuned to adapt to the task of classifying the input images into three classes: Healthy, DR, and Glaucoma. This strategy allows the model to gradually enhance its classification capabilities, which is beneficial in situations where there are only a limited number of labeled datasets available. Perturbation methods are also used to identify the input image characteristics responsible for influencing the models decision-making process. We achieve an overall accuracy of 91% on publicly available datasets.
Are Vision xLSTM Embedded UNet More Reliable in Medical 3D Image Segmentation?
Jun 24, 2024Abstract:The advancement of developing efficient medical image segmentation has evolved from initial dependence on Convolutional Neural Networks (CNNs) to the present investigation of hybrid models that combine CNNs with Vision Transformers. Furthermore, there is an increasing focus on creating architectures that are both high-performing in medical image segmentation tasks and computationally efficient to be deployed on systems with limited resources. Although transformers have several advantages like capturing global dependencies in the input data, they face challenges such as high computational and memory complexity. This paper investigates the integration of CNNs and Vision Extended Long Short-Term Memory (Vision-xLSTM) models by introducing a novel approach called UVixLSTM. The Vision-xLSTM blocks captures temporal and global relationships within the patches extracted from the CNN feature maps. The convolutional feature reconstruction path upsamples the output volume from the Vision-xLSTM blocks to produce the segmentation output. Our primary objective is to propose that Vision-xLSTM forms a reliable backbone for medical image segmentation tasks, offering excellent segmentation performance and reduced computational complexity. UVixLSTM exhibits superior performance compared to state-of-the-art networks on the publicly-available Synapse dataset. Code is available at: https://github.com/duttapallabi2907/UVixLSTM
Mixed Attention with Deep Supervision for Delineation of COVID Infection in Lung CT
Jan 17, 2023



Abstract:The COVID-19 pandemic, with its multiple variants, has placed immense pressure on the global healthcare system. An early effective screening and grading become imperative towards optimizing the limited available resources of the medical facilities. Computed tomography (CT) provides a significant non-invasive screening mechanism for COVID-19 infection. An automated segmentation of the infected volumes in lung CT is expected to significantly aid in the diagnosis and care of patients. However, an accurate demarcation of lesions remains problematic due to their irregular structure and location(s) within the lung. A novel deep learning architecture, Mixed Attention Deeply Supervised Network (MiADS-Net), is proposed for delineating the infected regions of the lung from CT images. Incorporating dilated convolutions with varying dilation rates, into a mixed attention framework, allows capture of multi-scale features towards improved segmentation of lesions having different sizes and textures. Mixed attention helps prioritise relevant feature maps to be probed, along with those regions containing crucial information within these maps. Deep supervision facilitates discovery of robust and discriminatory characteristics in the hidden layers at shallower levels, while overcoming the vanishing gradient. This is followed by estimating the severity of the disease, based on the ratio of the area of infected region in each lung with respect to its entire volume. Experimental results, on three publicly available datasets, indicate that the MiADS-Net outperforms several state-of-the-art architectures in the COVID-19 lesion segmentation task; particularly in defining structures involving complex geometries.
Collective Intelligent Strategy for Improved Segmentation of COVID-19 from CT
Dec 23, 2022Abstract:The devastation caused by the coronavirus pandemic makes it imperative to design automated techniques for a fast and accurate detection. We propose a novel non-invasive tool, using deep learning and imaging, for delineating COVID-19 infection in lungs. The Ensembling Attention-based Multi-scaled Convolution network (EAMC), employing Leave-One-Patient-Out (LOPO) training, exhibits high sensitivity and precision in outlining infected regions along with assessment of severity. The Attention module combines contextual with local information, at multiple scales, for accurate segmentation. Ensemble learning integrates heterogeneity of decision through different base classifiers. The superiority of EAMC, even with severe class imbalance, is established through comparison with existing state-of-the-art learning models over four publicly-available COVID-19 datasets. The results are suggestive of the relevance of deep learning in providing assistive intelligence to medical practitioners, when they are overburdened with patients as in pandemics. Its clinical significance lies in its unprecedented scope in providing low-cost decision-making for patients lacking specialized healthcare at remote locations.
Full-scale Deeply Supervised Attention Network for Segmenting COVID-19 Lesions
Oct 27, 2022Abstract:Automated delineation of COVID-19 lesions from lung CT scans aids the diagnosis and prognosis for patients. The asymmetric shapes and positioning of the infected regions make the task extremely difficult. Capturing information at multiple scales will assist in deciphering features, at global and local levels, to encompass lesions of variable size and texture. We introduce the Full-scale Deeply Supervised Attention Network (FuDSA-Net), for efficient segmentation of corona-infected lung areas in CT images. The model considers activation responses from all levels of the encoding path, encompassing multi-scalar features acquired at different levels of the network. This helps segment target regions (lesions) of varying shape, size and contrast. Incorporation of the entire gamut of multi-scalar characteristics into the novel attention mechanism helps prioritize the selection of activation responses and locations containing useful information. Determining robust and discriminatory features along the decoder path is facilitated with deep supervision. Connections in the decoder arm are remodeled to handle the issue of vanishing gradient. As observed from the experimental results, FuDSA-Net surpasses other state-of-the-art architectures; especially, when it comes to characterizing complicated geometries of the lesions.
Granular Generalized Variable Precision Rough Sets and Rational Approximations
May 31, 2022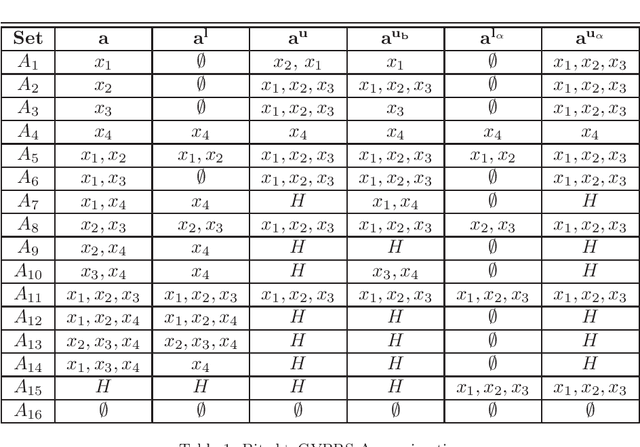
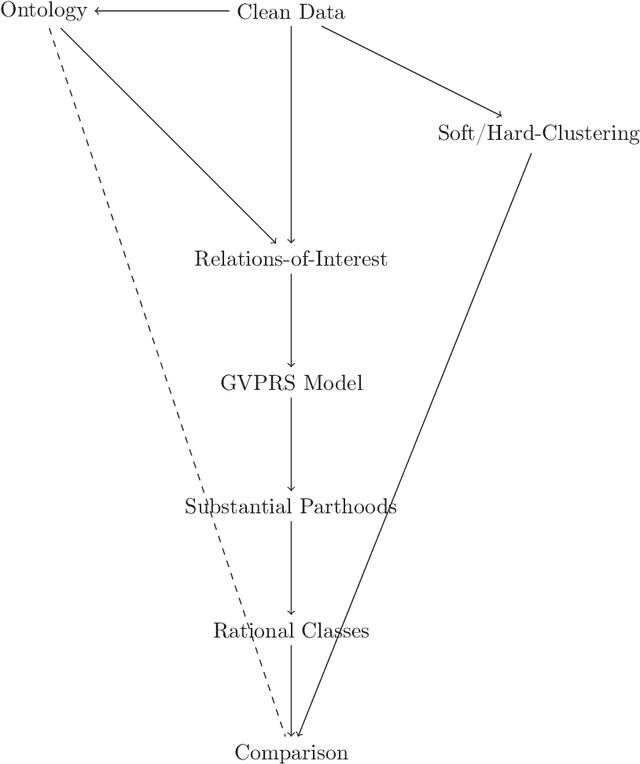
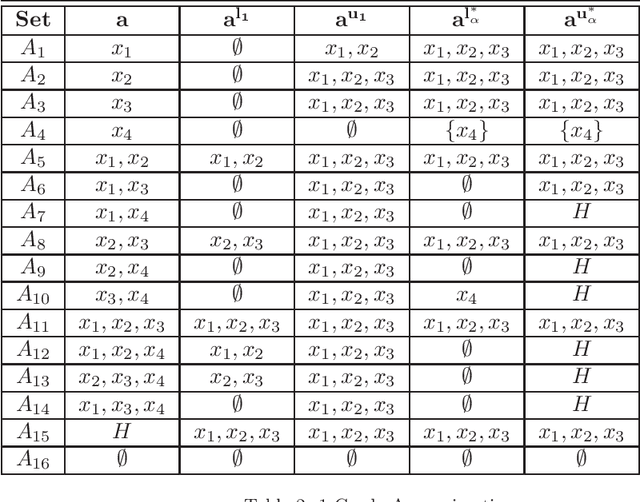
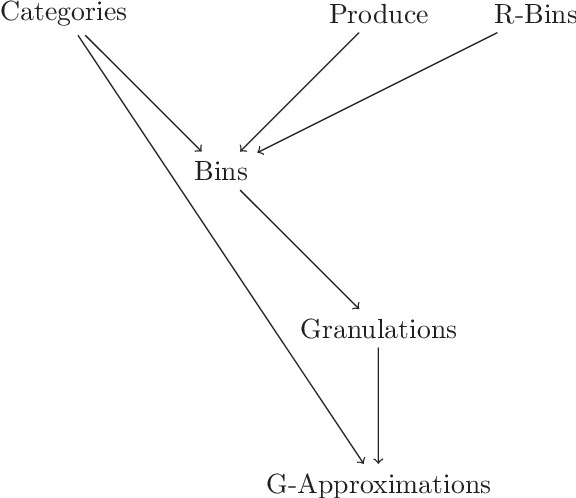
Abstract:Rational approximations are introduced and studied in granular graded sets and generalizations thereof by the first author in recent research papers. The concept of rationality is determined by related ontologies and coherence between granularity, parthood perspective and approximations used in the context. In addition, a framework is introduced by her in the mentioned paper(s). Granular approximations constructed as per the procedures of VPRS are likely to be more rational than those constructed from a classical perspective under certain conditions. This may continue to hold for some generalizations of the former; however, a formal characterization of such conditions is not available in the previously published literature. In this research, theoretical aspects of the problem are critically examined, uniform generalizations of granular VPRS are introduced, new connections with granular graded rough sets are proved, appropriate concepts of substantial parthood are introduced, and their extent of compatibility with the framework is accessed. Furthermore, meta applications to cluster validation, image segmentation and dynamic sorting are invented. Basic assumptions made are explained, and additional examples are constructed for readability.
Analysis of MRI Biomarkers for Brain Cancer Survival Prediction
Sep 03, 2021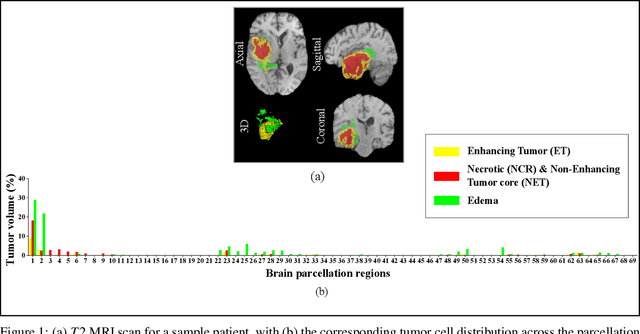
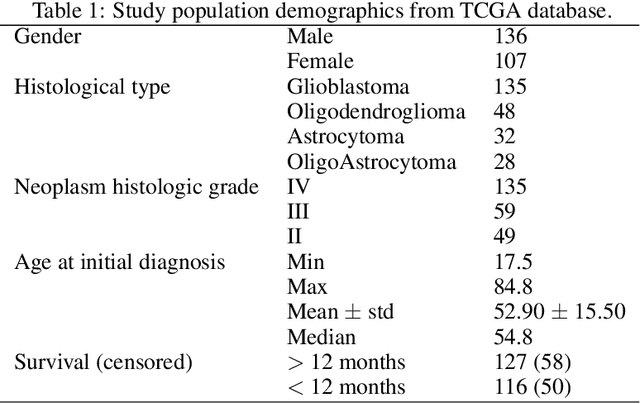
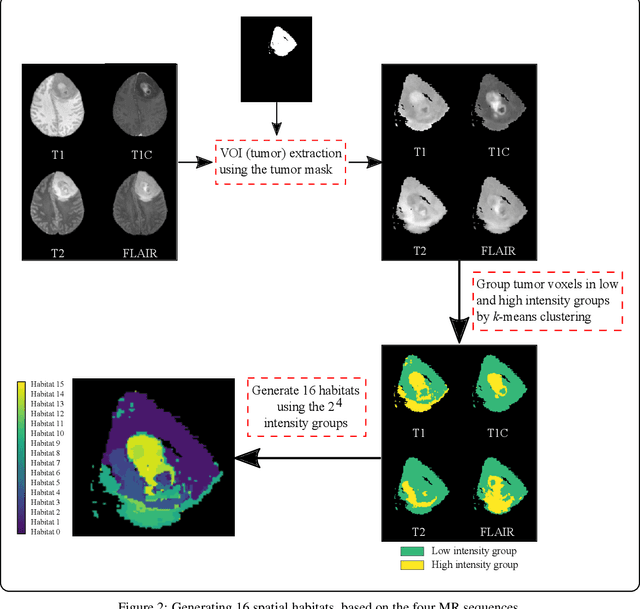
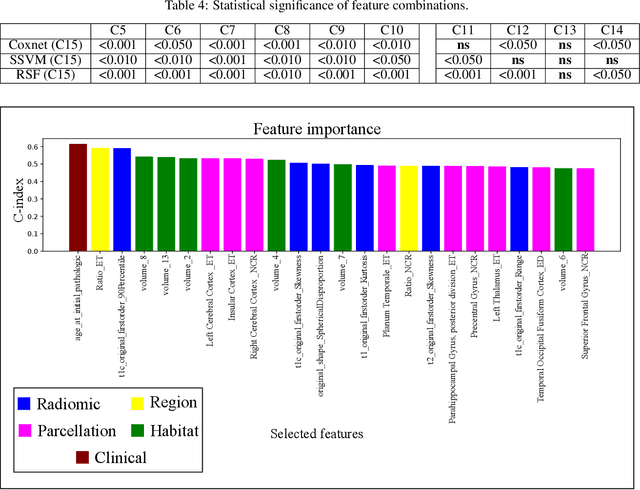
Abstract:Prediction of Overall Survival (OS) of brain cancer patients from multi-modal MRI is a challenging field of research. Most of the existing literature on survival prediction is based on Radiomic features, which does not consider either non-biological factors or the functional neurological status of the patient(s). Besides, the selection of an appropriate cut-off for survival and the presence of censored data create further problems. Application of deep learning models for OS prediction is also limited due to the lack of large annotated publicly available datasets. In this scenario we analyse the potential of two novel neuroimaging feature families, extracted from brain parcellation atlases and spatial habitats, along with classical radiomic and geometric features; to study their combined predictive power for analysing overall survival. A cross validation strategy with grid search is proposed to simultaneously select and evaluate the most predictive feature subset based on its predictive power. A Cox Proportional Hazard (CoxPH) model is employed for univariate feature selection, followed by the prediction of patient-specific survival functions by three multivariate parsimonious models viz. Coxnet, Random survival forests (RSF) and Survival SVM (SSVM). The brain cancer MRI data used for this research was taken from two open-access collections TCGA-GBM and TCGA-LGG available from The Cancer Imaging Archive (TCIA). Corresponding survival data for each patient was downloaded from The Cancer Genome Atlas (TCGA). A high cross validation $C-index$ score of $0.82\pm.10$ was achieved using RSF with the best $24$ selected features. Age was found to be the most important biological predictor. There were $9$, $6$, $6$ and $2$ features selected from the parcellation, habitat, radiomic and region-based feature groups respectively.
Deep Learning for Screening COVID-19 using Chest X-Ray Images
Apr 24, 2020

Abstract:With the ever increasing demand for screening millions of prospective "novel coronavirus" or COVID-19 cases, and due to the emergence of high false negatives in the commonly used PCR tests, the necessity for probing an alternative simple screening mechanism of COVID-19 using radiological images (like chest X-Rays) assumes importance. In this scenario, machine learning (ML) and deep learning (DL) offer fast, automated, effective strategies to detect abnormalities and extract key features of the altered lung parenchyma, which may be related to specific signatures of the COVID-19 virus. However, the available COVID-19 datasets are inadequate to train deep neural networks. Therefore, we propose a new concept called domain extension transfer learning (DETL). We employ DETL, with pre-trained deep convolutional neural network, on a related large chest X-Ray dataset that is tuned for classifying between four classes viz. $normal$, $other\_disease$, $pneumonia$ and $Covid-19$. A 5-fold cross validation is performed to estimate the feasibility of using chest X-Rays to diagnose COVID-19. The initial results show promise, with the possibility of replication on bigger and more diverse data sets. The overall accuracy was measured as $95.3\% \pm 0.02$. In order to get an idea about the COVID-19 detection transparency, we employed the concept of Gradient Class Activation Map (Grad-CAM) for detecting the regions where the model paid more attention during the classification. This was found to strongly correlate with clinical findings, as validated by experts.
Deep Radiomics for Brain Tumor Detection and Classification from Multi-Sequence MRI
Mar 21, 2019
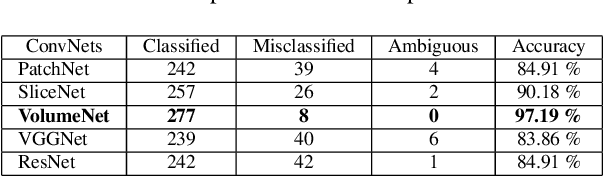


Abstract:Glioma constitutes 80% of malignant primary brain tumors and is usually classified as HGG and LGG. The LGG tumors are less aggressive, with slower growth rate as compared to HGG, and are responsive to therapy. Tumor biopsy being challenging for brain tumor patients, noninvasive imaging techniques like Magnetic Resonance Imaging (MRI) have been extensively employed in diagnosing brain tumors. Therefore automated systems for the detection and prediction of the grade of tumors based on MRI data becomes necessary for assisting doctors in the framework of augmented intelligence. In this paper, we thoroughly investigate the power of Deep ConvNets for classification of brain tumors using multi-sequence MR images. We propose novel ConvNet models, which are trained from scratch, on MRI patches, slices, and multi-planar volumetric slices. The suitability of transfer learning for the task is next studied by applying two existing ConvNets models (VGGNet and ResNet) trained on ImageNet dataset, through fine-tuning of the last few layers. LOPO testing, and testing on the holdout dataset are used to evaluate the performance of the ConvNets. Results demonstrate that the proposed ConvNets achieve better accuracy in all cases where the model is trained on the multi-planar volumetric dataset. Unlike conventional models, it obtains a testing accuracy of 95% for the low/high grade glioma classification problem. A score of 97% is generated for classification of LGG with/without 1p/19q codeletion, without any additional effort towards extraction and selection of features. We study the properties of self-learned kernels/ filters in different layers, through visualization of the intermediate layer outputs. We also compare the results with that of state-of-the-art methods, demonstrating a maximum improvement of 7% on the grading performance of ConvNets and 9% on the prediction of 1p/19q codeletion status.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge