Eptehal Nashnoush
Tuning In: Analysis of Audio Classifier Performance in Clinical Settings with Limited Data
Feb 19, 2024Abstract:This study assesses deep learning models for audio classification in a clinical setting with the constraint of small datasets reflecting real-world prospective data collection. We analyze CNNs, including DenseNet and ConvNeXt, alongside transformer models like ViT, SWIN, and AST, and compare them against pre-trained audio models such as YAMNet and VGGish. Our method highlights the benefits of pre-training on large datasets before fine-tuning on specific clinical data. We prospectively collected two first-of-their-kind patient audio datasets from stroke patients. We investigated various preprocessing techniques, finding that RGB and grayscale spectrogram transformations affect model performance differently based on the priors they learn from pre-training. Our findings indicate CNNs can match or exceed transformer models in small dataset contexts, with DenseNet-Contrastive and AST models showing notable performance. This study highlights the significance of incremental marginal gains through model selection, pre-training, and preprocessing in sound classification; this offers valuable insights for clinical diagnostics that rely on audio classification.
Can Self Reported Symptoms Predict Daily COVID-19 Cases?
May 18, 2021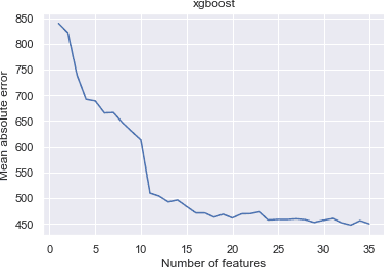
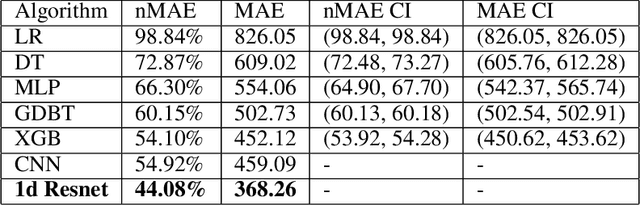
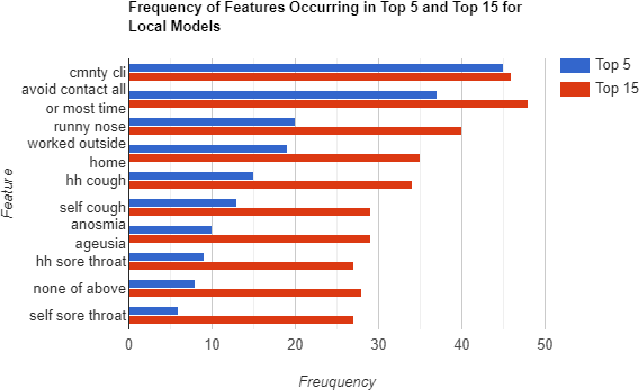

Abstract:The COVID-19 pandemic has impacted lives and economies across the globe, leading to many deaths. While vaccination is an important intervention, its roll-out is slow and unequal across the globe. Therefore, extensive testing still remains one of the key methods to monitor and contain the virus. Testing on a large scale is expensive and arduous. Hence, we need alternate methods to estimate the number of cases. Online surveys have been shown to be an effective method for data collection amidst the pandemic. In this work, we develop machine learning models to estimate the prevalence of COVID-19 using self-reported symptoms. Our best model predicts the daily cases with a mean absolute error (MAE) of 226.30 (normalized MAE of 27.09%) per state, which demonstrates the possibility of predicting the actual number of confirmed cases by utilizing self-reported symptoms. The models are developed at two levels of data granularity - local models, which are trained at the state level, and a single global model which is trained on the combined data aggregated across all states. Our results indicate a lower error on the local models as opposed to the global model. In addition, we also show that the most important symptoms (features) vary considerably from state to state. This work demonstrates that the models developed on crowd-sourced data, curated via online platforms, can complement the existing epidemiological surveillance infrastructure in a cost-effective manner.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge