Dongkyun Kim
CXR-LT 2024: A MICCAI challenge on long-tailed, multi-label, and zero-shot disease classification from chest X-ray
Jun 09, 2025Abstract:The CXR-LT series is a community-driven initiative designed to enhance lung disease classification using chest X-rays (CXR). It tackles challenges in open long-tailed lung disease classification and enhances the measurability of state-of-the-art techniques. The first event, CXR-LT 2023, aimed to achieve these goals by providing high-quality benchmark CXR data for model development and conducting comprehensive evaluations to identify ongoing issues impacting lung disease classification performance. Building on the success of CXR-LT 2023, the CXR-LT 2024 expands the dataset to 377,110 chest X-rays (CXRs) and 45 disease labels, including 19 new rare disease findings. It also introduces a new focus on zero-shot learning to address limitations identified in the previous event. Specifically, CXR-LT 2024 features three tasks: (i) long-tailed classification on a large, noisy test set, (ii) long-tailed classification on a manually annotated "gold standard" subset, and (iii) zero-shot generalization to five previously unseen disease findings. This paper provides an overview of CXR-LT 2024, detailing the data curation process and consolidating state-of-the-art solutions, including the use of multimodal models for rare disease detection, advanced generative approaches to handle noisy labels, and zero-shot learning strategies for unseen diseases. Additionally, the expanded dataset enhances disease coverage to better represent real-world clinical settings, offering a valuable resource for future research. By synthesizing the insights and innovations of participating teams, we aim to advance the development of clinically realistic and generalizable diagnostic models for chest radiography.
Attribute Based Interpretable Evaluation Metrics for Generative Models
Oct 26, 2023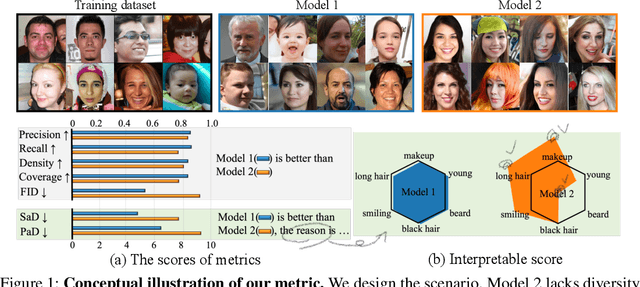
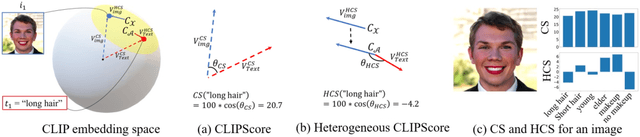
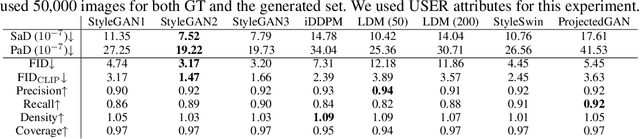
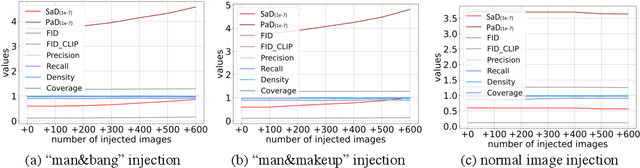
Abstract:When the training dataset comprises a 1:1 proportion of dogs to cats, a generative model that produces 1:1 dogs and cats better resembles the training species distribution than another model with 3:1 dogs and cats. Can we capture this phenomenon using existing metrics? Unfortunately, we cannot, because these metrics do not provide any interpretability beyond "diversity". In this context, we propose a new evaluation protocol that measures the divergence of a set of generated images from the training set regarding the distribution of attribute strengths as follows. Single-attribute Divergence (SaD) measures the divergence regarding PDFs of a single attribute. Paired-attribute Divergence (PaD) measures the divergence regarding joint PDFs of a pair of attributes. They provide which attributes the models struggle. For measuring the attribute strengths of an image, we propose Heterogeneous CLIPScore (HCS) which measures the cosine similarity between image and text vectors with heterogeneous initial points. With SaD and PaD, we reveal the following about existing generative models. ProjectedGAN generates implausible attribute relationships such as a baby with a beard even though it has competitive scores of existing metrics. Diffusion models struggle to capture diverse colors in the datasets. The larger sampling timesteps of latent diffusion model generate the more minor objects including earrings and necklaces. Stable Diffusion v1.5 better captures the attributes than v2.1. Our metrics lay a foundation for explainable evaluations of generative models.
Towards long-tailed, multi-label disease classification from chest X-ray: Overview of the CXR-LT challenge
Oct 24, 2023



Abstract:Many real-world image recognition problems, such as diagnostic medical imaging exams, are "long-tailed" $\unicode{x2013}$ there are a few common findings followed by many more relatively rare conditions. In chest radiography, diagnosis is both a long-tailed and multi-label problem, as patients often present with multiple findings simultaneously. While researchers have begun to study the problem of long-tailed learning in medical image recognition, few have studied the interaction of label imbalance and label co-occurrence posed by long-tailed, multi-label disease classification. To engage with the research community on this emerging topic, we conducted an open challenge, CXR-LT, on long-tailed, multi-label thorax disease classification from chest X-rays (CXRs). We publicly release a large-scale benchmark dataset of over 350,000 CXRs, each labeled with at least one of 26 clinical findings following a long-tailed distribution. We synthesize common themes of top-performing solutions, providing practical recommendations for long-tailed, multi-label medical image classification. Finally, we use these insights to propose a path forward involving vision-language foundation models for few- and zero-shot disease classification.
CheXFusion: Effective Fusion of Multi-View Features using Transformers for Long-Tailed Chest X-Ray Classification
Aug 08, 2023Abstract:Medical image classification poses unique challenges due to the long-tailed distribution of diseases, the co-occurrence of diagnostic findings, and the multiple views available for each study or patient. This paper introduces our solution to the ICCV CVAMD 2023 Shared Task on CXR-LT: Multi-Label Long-Tailed Classification on Chest X-Rays. Our approach introduces CheXFusion, a transformer-based fusion module incorporating multi-view images. The fusion module, guided by self-attention and cross-attention mechanisms, efficiently aggregates multi-view features while considering label co-occurrence. Furthermore, we explore data balancing and self-training methods to optimize the model's performance. Our solution achieves state-of-the-art results with 0.372 mAP in the MIMIC-CXR test set, securing 1st place in the competition. Our success in the task underscores the significance of considering multi-view settings, class imbalance, and label co-occurrence in medical image classification. Public code is available at https://github.com/dongkyuk/CXR-LT-public-solution
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge