Diego Sona
AIforCOVID: predicting the clinical outcomes in patients with COVID-19 applying AI to chest-X-rays. An Italian multicentre study
Dec 11, 2020
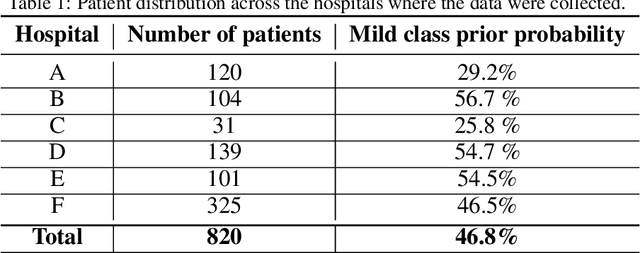
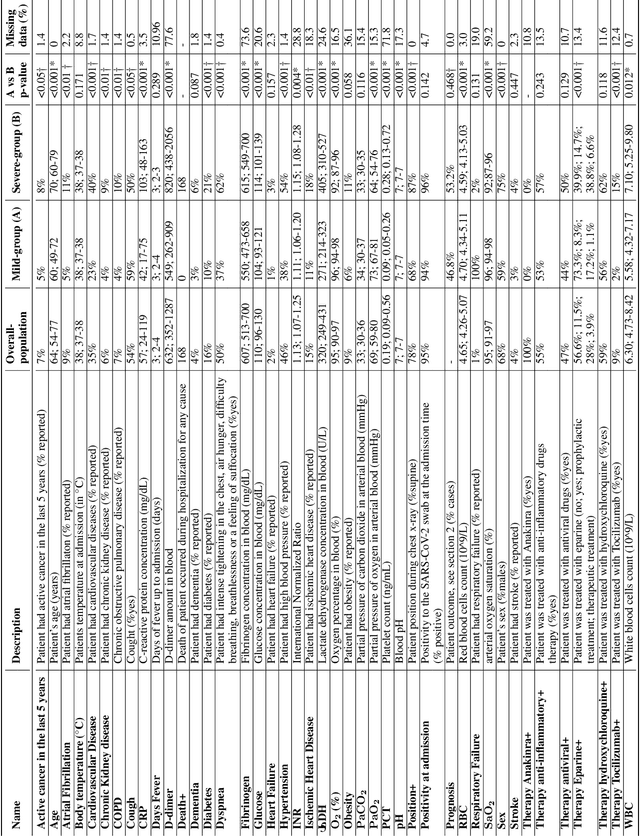
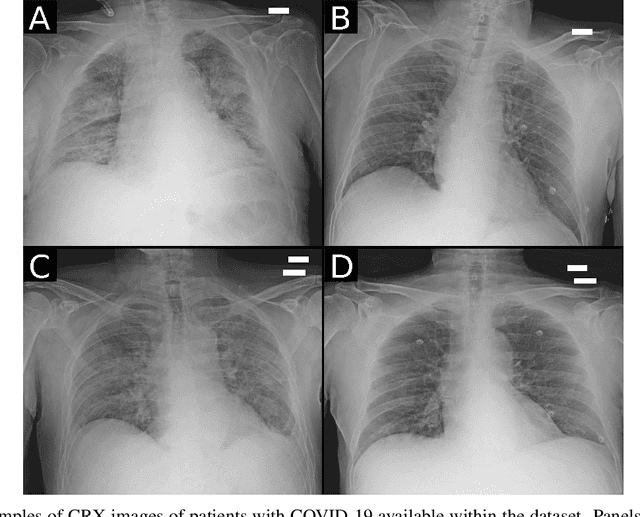
Abstract:Recent epidemiological data report that worldwide more than 53 million people have been infected by SARS-CoV-2, resulting in 1.3 million deaths. The disease has been spreading very rapidly and few months after the identification of the first infected, shortage of hospital resources quickly became a problem. In this work we investigate whether chest X-ray (CXR) can be used as a possible tool for the early identification of patients at risk of severe outcome, like intensive care or death. CXR is a radiological technique that compared to computed tomography (CT) it is simpler, faster, more widespread and it induces lower radiation dose. We present a dataset including data collected from 820 patients by six Italian hospitals in spring 2020 during the first COVID-19 emergency. The dataset includes CXR images, several clinical attributes and clinical outcomes. We investigate the potential of artificial intelligence to predict the prognosis of such patients, distinguishing between severe and mild cases, thus offering a baseline reference for other researchers and practitioners. To this goal, we present three approaches that use features extracted from CXR images, either handcrafted or automatically by convolutional neuronal networks, which are then integrated with the clinical data. Exhaustive evaluation shows promising performance both in 10-fold and leave-one-centre-out cross-validation, implying that clinical data and images have the potential to provide useful information for the management of patients and hospital resources.
Weakly Supervised Geodesic Segmentation of Egyptian Mummy CT Scans
Apr 17, 2020



Abstract:In this paper, we tackle the task of automatically analyzing 3D volumetric scans obtained from computed tomography (CT) devices. In particular, we address a particular task for which data is very limited: the segmentation of ancient Egyptian mummies CT scans. We aim at digitally unwrapping the mummy and identify different segments such as body, bandages and jewelry. The problem is complex because of the lack of annotated data for the different semantic regions to segment, thus discouraging the use of strongly supervised approaches. We, therefore, propose a weakly supervised and efficient interactive segmentation method to solve this challenging problem. After segmenting the wrapped mummy from its exterior region using histogram analysis and template matching, we first design a voxel distance measure to find an approximate solution for the body and bandage segments. Here, we use geodesic distances since voxel features as well as spatial relationship among voxels is incorporated in this measure. Next, we refine the solution using a GrabCut based segmentation together with a tracking method on the slices of the scan that assigns labels to different regions in the volume, using limited supervision in the form of scribbles drawn by the user. The efficiency of the proposed method is demonstrated using visualizations and validated through quantitative measures and qualitative unwrapping of the mummy.
Scanner Invariant Multiple Sclerosis Lesion Segmentation from MRI
Oct 22, 2019



Abstract:This paper presents a simple and effective generalization method for magnetic resonance imaging (MRI) segmentation when data is collected from multiple MRI scanning sites and as a consequence is affected by (site-)domain shifts. We propose to integrate a traditional encoder-decoder network with a regularization network. This added network includes an auxiliary loss term which is responsible for the reduction of the domain shift problem and for the resulting improved generalization. The proposed method was evaluated on multiple sclerosis lesion segmentation from MRI data. We tested the proposed model on an in-house clinical dataset including 117 patients from 56 different scanning sites. In the experiments, our method showed better generalization performance than other baseline networks.
Multi-branch Convolutional Neural Network for Multiple Sclerosis Lesion Segmentation
Nov 16, 2018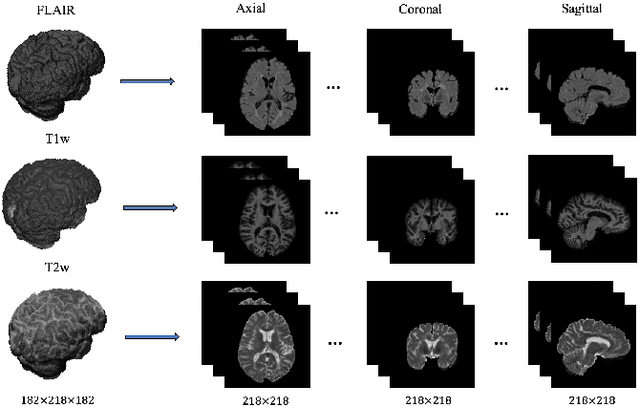
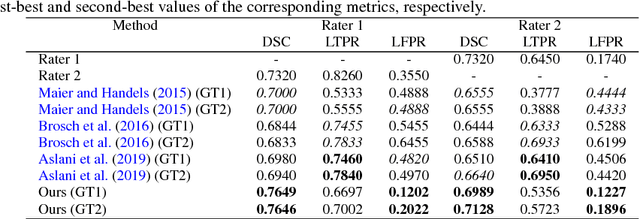
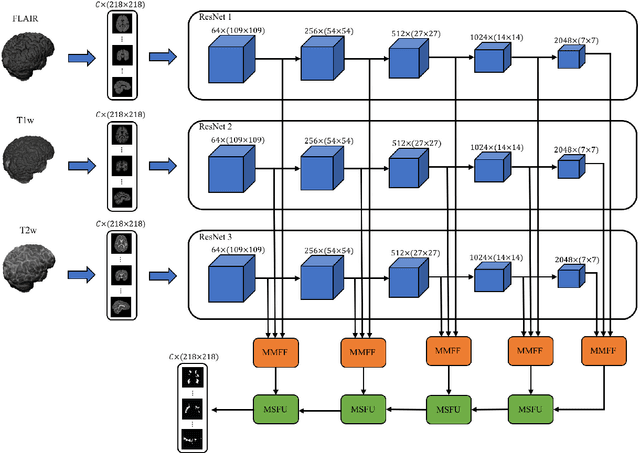
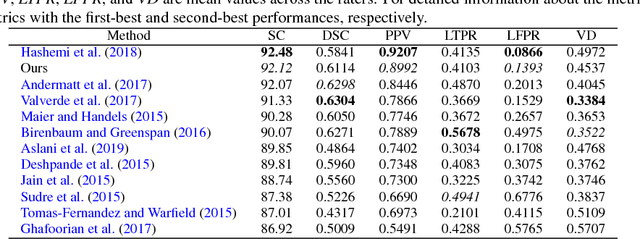
Abstract:In this paper, we present an automated approach for segmenting multiple sclerosis (MS) lesions from multi-modal brain magnetic resonance images. Our method is based on a deep end-to-end 2D convolutional neural network (CNN) for slice-based segmentation of 3D volumetric data. The proposed CNN includes a multi-branch downsampling path, which enables the network to encode slices from multiple modalities separately. Multi-scale feature fusion blocks are proposed to combine feature maps from different modalities at different stages of the network. Then, multi-scale feature upsampling blocks are introduced to upsize combined feature maps with different resolutions to leverage information from the lesion shape and location. We trained and tested our model using orthogonal plane orientations of each 3D modality to exploit the contextual information in all directions. The proposed pipeline is evaluated on two different datasets: a private dataset including 37 MS patients and a publicly available dataset known as the ISBI 2015 longitudinal MS lesion segmentation challenge dataset, consisting of 14 MS patients. Considering the ISBI challenge, at the time of submission, our method was amongst the top performing solutions. On the private dataset, using the same array of performance metrics as in the ISBI challenge, the proposed approach shows high improvements in MS lesion segmentation comparing with other publicly available tools.
Modeling Retinal Ganglion Cell Population Activity with Restricted Boltzmann Machines
Jan 17, 2017



Abstract:The retina is a complex nervous system which encodes visual stimuli before higher order processing occurs in the visual cortex. In this study we evaluated whether information about the stimuli received by the retina can be retrieved from the firing rate distribution of Retinal Ganglion Cells (RGCs), exploiting High-Density 64x64 MEA technology. To this end, we modeled the RGC population activity using mean-covariance Restricted Boltzmann Machines, latent variable models capable of learning the joint distribution of a set of continuous observed random variables and a set of binary unobserved random units. The idea was to figure out if binary latent states encode the regularities associated to different visual stimuli, as modes in the joint distribution. We measured the goodness of mcRBM encoding by calculating the Mutual Information between the latent states and the stimuli shown to the retina. Results show that binary states can encode the regularities associated to different stimuli, using both gratings and natural scenes as stimuli. We also discovered that hidden variables encode interesting properties of retinal activity, interpreted as population receptive fields. We further investigated the ability of the model to learn different modes in population activity by comparing results associated to a retina in normal conditions and after pharmacologically blocking GABA receptors (GABAC at first, and then also GABAA and GABAB). As expected, Mutual Information tends to decrease if we pharmacologically block receptors. We finally stress that the computational method described in this work could potentially be applied to any kind of neural data obtained through MEA technology, though different techniques should be applied to interpret the results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge