Michael Dayan
Scanner Invariant Multiple Sclerosis Lesion Segmentation from MRI
Oct 22, 2019



Abstract:This paper presents a simple and effective generalization method for magnetic resonance imaging (MRI) segmentation when data is collected from multiple MRI scanning sites and as a consequence is affected by (site-)domain shifts. We propose to integrate a traditional encoder-decoder network with a regularization network. This added network includes an auxiliary loss term which is responsible for the reduction of the domain shift problem and for the resulting improved generalization. The proposed method was evaluated on multiple sclerosis lesion segmentation from MRI data. We tested the proposed model on an in-house clinical dataset including 117 patients from 56 different scanning sites. In the experiments, our method showed better generalization performance than other baseline networks.
Multi-branch Convolutional Neural Network for Multiple Sclerosis Lesion Segmentation
Nov 16, 2018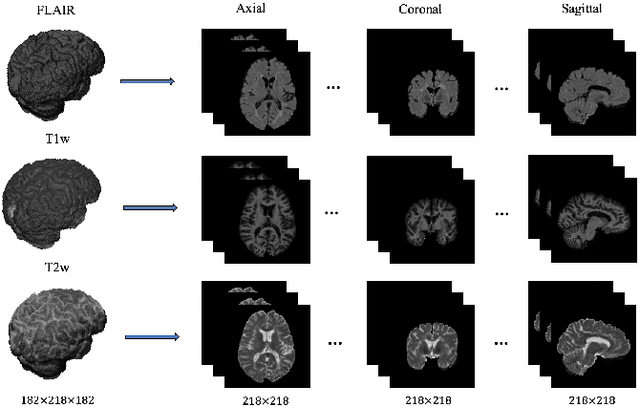
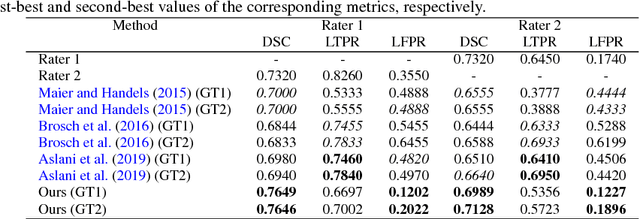
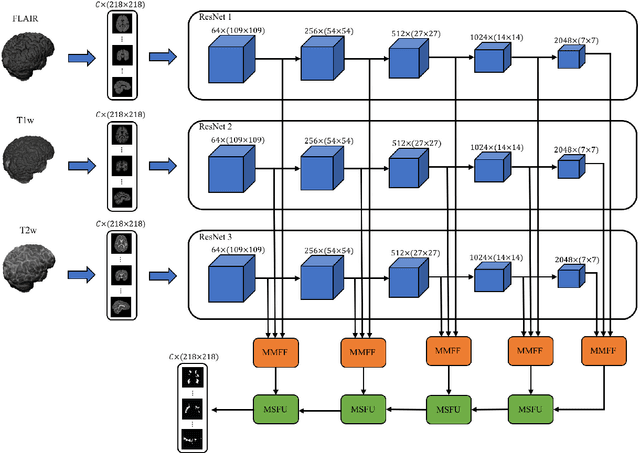
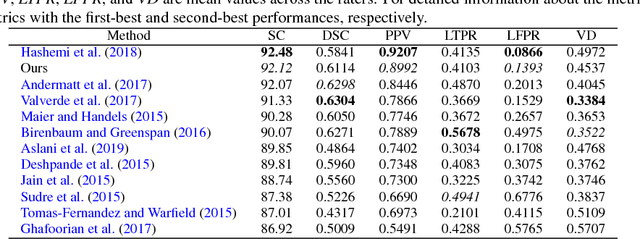
Abstract:In this paper, we present an automated approach for segmenting multiple sclerosis (MS) lesions from multi-modal brain magnetic resonance images. Our method is based on a deep end-to-end 2D convolutional neural network (CNN) for slice-based segmentation of 3D volumetric data. The proposed CNN includes a multi-branch downsampling path, which enables the network to encode slices from multiple modalities separately. Multi-scale feature fusion blocks are proposed to combine feature maps from different modalities at different stages of the network. Then, multi-scale feature upsampling blocks are introduced to upsize combined feature maps with different resolutions to leverage information from the lesion shape and location. We trained and tested our model using orthogonal plane orientations of each 3D modality to exploit the contextual information in all directions. The proposed pipeline is evaluated on two different datasets: a private dataset including 37 MS patients and a publicly available dataset known as the ISBI 2015 longitudinal MS lesion segmentation challenge dataset, consisting of 14 MS patients. Considering the ISBI challenge, at the time of submission, our method was amongst the top performing solutions. On the private dataset, using the same array of performance metrics as in the ISBI challenge, the proposed approach shows high improvements in MS lesion segmentation comparing with other publicly available tools.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge